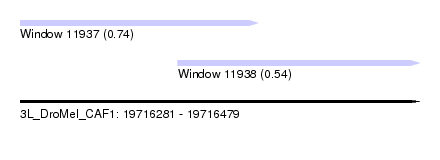
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,716,281 – 19,716,479 |
| Length | 198 |
| Max. P | 0.741713 |

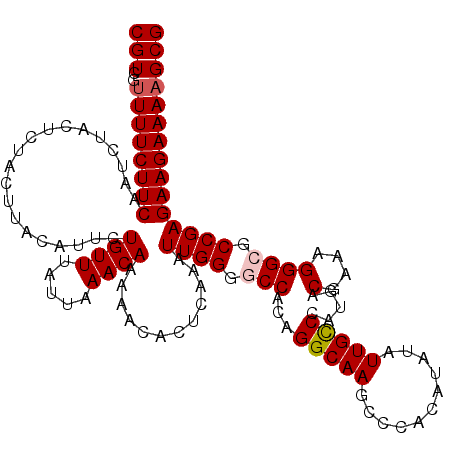
| Location | 19,716,281 – 19,716,399 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19716281 118 + 23771897 CGUCGUUUUCUUCAAUCUACUCUACUUACAUUCUGUUUAUUAAACAAAAACACUCAAAUUGGGGCCACAGGCAAGCCC--AUAUAUUGCCCAUGACGAAAGGGCGCCGAGAAGAAAAGCG (((..((((((((....................((((.....))))............((((.(((...(((((....--.....))))).....(....)))).))))))))))))))) ( -28.10) >DroSec_CAF1 25431 120 + 1 CGUCGUUUUCUUCAAUCUACUCUACUUACAUUCUGUUUAUUAAACAAAAACACUCAAAUUGGGGCCACAGGCAAGCCCACAUAUAUUGCCCAUAACGAAAGGGCGCCGAGAAGAAAAGCG (((..((((((((....................((((.....))))............((((.(((...(((((...........))))).....(....)))).))))))))))))))) ( -27.90) >DroSim_CAF1 27656 120 + 1 CGUCGUUUUCUUCAAUCUACUCUACUUACAUUCUGUUUAUUAAACAAAAACACUCAAAUUGGGGCCACAGGCAAGCCCACAUAUAUUGCCCAUGACGAAAGGGCGCCGAGAAGAAAAGCG (((..((((((((....................((((.....))))............((((.(((...(((((...........))))).....(....)))).))))))))))))))) ( -27.90) >DroEre_CAF1 28181 117 + 1 CGUCAUUUUCUUCAAUCUACUCUACUUACAUUCUGUUUAUUAAACAAAAACACUCAAAUUGGGACCGCAGGCAAGCCCAC--AUAUUGCCCAUGACGA-AGGGCCCCGAGAAGAAAAGCG (((..((((((((....................((((.....))))............(((((......((....))...--.....((((.......-.)))))))))))))))))))) ( -23.70) >DroYak_CAF1 26378 117 + 1 CGUCGUUUUCUUCAAUCUACUCUACUUACAUUCUGUUUAUUAAACAAAAACACUCAAAUUGGGACCACAGGCAAGCCCAC--AUAUUGUCCAUGACGA-AGGGCCCCGAGAAGAAACGCG ((.(((((.((((....................((((.....))))............(((((.((...((....))...--...(((((...)))))-..)).)))))))))))))))) ( -23.20) >consensus CGUCGUUUUCUUCAAUCUACUCUACUUACAUUCUGUUUAUUAAACAAAAACACUCAAAUUGGGGCCACAGGCAAGCCCACAUAUAUUGCCCAUGACGAAAGGGCGCCGAGAAGAAAAGCG (((..((((((((....................((((.....))))............((((.(((...(((((...........))))).....(....)))).))))))))))))))) (-23.06 = -23.50 + 0.44)

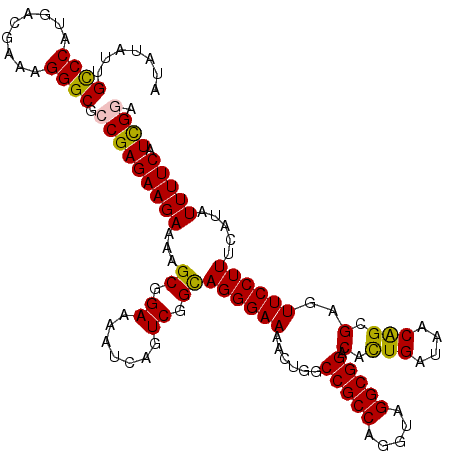
| Location | 19,716,359 – 19,716,479 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -29.76 |
| Energy contribution | -29.38 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19716359 120 + 23771897 AUAUAUUGCCCAUGACGAAAGGGCGCCGAGAAGAAAAGCGGAAAAUCAGUCGGCAGGGAAAACUGGCCGCCAGGUAGGCGGACACUGAUAACAGCGAUUUCCUUUCAUAUUUUCAUCGGA .......((((.........)))).(((((((((...((.((.......)).))(((((((.....(((((.....))))).(.(((....))).).))))))).....))))).)))). ( -36.60) >DroSec_CAF1 25511 120 + 1 AUAUAUUGCCCAUAACGAAAGGGCGCCGAGAAGAAAAGCGGAAAAUCAGUCGGCAGGGAAAACUGGCCGCCAGGUAGGCGGACAUUGAUAACAGCGAGUUCCUUUCAUAUUUUCAUCGGA .......((((.........)))).(((((((((...((.((.......)).))((((((..(((.(((((.....)))))..........)))....)))))).....))))).)))). ( -34.90) >DroSim_CAF1 27736 108 + 1 AUAUAUUGCCCAUGACGAAAGGGCGCCGAGAAGAAAAGCG------------GCAGGGAAAACUGGCCGCCAGGUAGGCGGACACUGAUAACAGCGAGUUCCUUUCAUAUUUUCAUCGGA .......((((.........)))).((((...(((((...------------(.((((((......(((((.....))))).(.(((....))).)..)))))).)...))))).)))). ( -33.90) >DroEre_CAF1 28261 117 + 1 --AUAUUGCCCAUGACGA-AGGGCCCCGAGAAGAAAAGCGGAAAAUAUGUCGGUAGGGAAAACUGCCCGCCAGGUAGGCGGACACUGAUAACGGAGAGUUCCUUUCAUAUUUUCAUCGAA --.....((((.......-.))))..(((...(((((...((((...(((((((((......)))))((((.....))))))))........((......))))))...))))).))).. ( -34.40) >DroYak_CAF1 26458 117 + 1 --AUAUUGUCCAUGACGA-AGGGCCCCGAGAAGAAACGCGGAAAAUCUGUCGGUAGGGAAAACUGCCCGCCAGGUAGGCGGACACUGAUAACAGCGAGUUCCUUUCAUAUUUUCAUUGAA --.....((((.......-.))))...((((.(((.(((.....((((((((((((......)))))((((.....))))))))..)))....)))..))).)))).............. ( -31.00) >consensus AUAUAUUGCCCAUGACGAAAGGGCGCCGAGAAGAAAAGCGGAAAAUCAGUCGGCAGGGAAAACUGGCCGCCAGGUAGGCGGACACUGAUAACAGCGAGUUCCUUUCAUAUUUUCAUCGGA .......((((.........)))).(((((((((...((.((.......)).))((((((......(((((.....))))).(.(((....))).)..)))))).....))))).)))). (-29.76 = -29.38 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:41 2006