| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,713,854 – 19,713,946 |
| Length | 92 |
| Max. P | 0.867325 |

| Location | 19,713,854 – 19,713,946 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.35 |
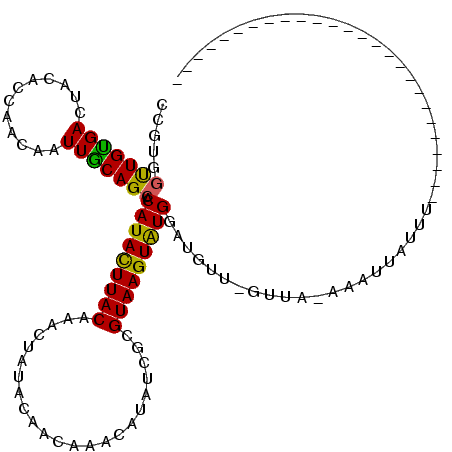
| Mean single sequence MFE | -20.83 |
| Consensus MFE | -15.27 |
| Energy contribution | -16.40 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19713854 92 + 23771897 --------------------------ACAUAAUUA-AAAA-AACAUCCAUACUUACGCGAUAUGUUUGUUGUAUAGUGUGUAAGUAUGUGCUGCAAUUGUUGGUGUAGUCGCAGCCACAG --------------------------.........-....-......((((((((((((.((((.......)))).)))))))))))).(((((.((((......)))).)))))..... ( -24.30) >DroSec_CAF1 23098 93 + 1 --------------------------AAAUAAUUU-AAAAUAUCAUUCAUACUUACGCGAUAUGUUUGUUGUAUAGUGUGUAAGUAUGUGCUGCAAUUGUUGGUGUAGUCACAACCACGG --------------------------.........-............(((((((((((.((((.......)))).)))))))))))((((((((........))))).)))........ ( -21.10) >DroEre_CAF1 25808 118 + 1 UUAUCUACUAAAUAUGCCCUAUUUAUAAACAUUAUAUAAC-UAAAUCCACACUUACGCGAUAUGUUUGUGGUAUAGUUUGUAAGUAUGUGCUGCAAUUGUUGGUGUAGUCACAA-CACGG .................(((((((((((((.(((......-)))..(((((...(((.....))).)))))....)))))))))))(((((((((........))))).)))).-...)) ( -21.70) >DroYak_CAF1 23909 118 + 1 -UUUAUAAUGUAAAUCCCCUAAAUAUAAACAAUAUAUAAC-AAAAUCCAUACUUACGCGAUAUGUUUGUUGUAUAGUUCGUAAAUAUGUGCUGUAAUUAUUUGCCUAGUCACAACCACAA -.......(((............((..(((..((((((((-(((...(((((......).))))))))))))))))))..))....((((((((((....))))..)).))))...))). ( -16.20) >consensus __________________________AAACAAUAU_AAAA_AAAAUCCAUACUUACGCGAUAUGUUUGUUGUAUAGUGUGUAAGUAUGUGCUGCAAUUGUUGGUGUAGUCACAACCACAG ................................................(((((((((((.((((.......)))).)))))))))))((((((((........))))).)))........ (-15.27 = -16.40 + 1.13)

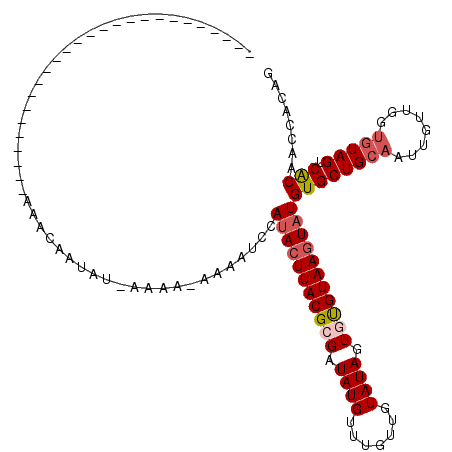
| Location | 19,713,854 – 19,713,946 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.35 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -13.41 |
| Energy contribution | -12.72 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19713854 92 - 23771897 CUGUGGCUGCGACUACACCAACAAUUGCAGCACAUACUUACACACUAUACAACAAACAUAUCGCGUAAGUAUGGAUGUU-UUUU-UAAUUAUGU-------------------------- .....(((((((............))))))).(((((((((.(...................).)))))))))......-....-.........-------------------------- ( -18.51) >DroSec_CAF1 23098 93 - 1 CCGUGGUUGUGACUACACCAACAAUUGCAGCACAUACUUACACACUAUACAACAAACAUAUCGCGUAAGUAUGAAUGAUAUUUU-AAAUUAUUU-------------------------- ..(..(((((..........)))))..)....(((((((((.(...................).)))))))))...........-.........-------------------------- ( -16.91) >DroEre_CAF1 25808 118 - 1 CCGUG-UUGUGACUACACCAACAAUUGCAGCACAUACUUACAAACUAUACCACAAACAUAUCGCGUAAGUGUGGAUUUA-GUUAUAUAAUGUUUAUAAAUAGGGCAUAUUUAGUAGAUAA ..(((-(((..(............)..)))))).........(((((..(((((.((.......))...)))))...))-)))......((((((((((((.....))))).))))))). ( -22.50) >DroYak_CAF1 23909 118 - 1 UUGUGGUUGUGACUAGGCAAAUAAUUACAGCACAUAUUUACGAACUAUACAACAAACAUAUCGCGUAAGUAUGGAUUUU-GUUAUAUAUUGUUUAUAUUUAGGGGAUUUACAUUAUAAA- .(((((((((((.((......)).))))))).)))).......................(((.(.(((((((((((..(-((...)))..))))))))))).).)))............- ( -21.50) >consensus CCGUGGUUGUGACUACACCAACAAUUGCAGCACAUACUUACAAACUAUACAACAAACAUAUCGCGUAAGUAUGGAUGUU_GUUA_AAAUUAUUU__________________________ .....(((((((............))))))).(((((((((.......................)))))))))............................................... (-13.41 = -12.72 + -0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:37 2006