
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,703,747 – 19,703,882 |
| Length | 135 |
| Max. P | 0.961577 |

| Location | 19,703,747 – 19,703,853 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.55 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -21.74 |
| Energy contribution | -23.26 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19703747 106 - 23771897 UACAUAUU--------UAUGUAGUUAU------AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAGCCGCGGCUUUGGCAUGUGUAGACACUAAACUUAUUAAUAA (((((...--------.))))).(..(------(((((((.....))))))))..).......((((((..((.(((((((((......)))).)))))))..))))))........... ( -26.60) >DroSec_CAF1 17824 109 - 1 UACAUACGUA-----GUAUGUAGUUAU------AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAGCCGUGGCUUUGGCAUGUGUAGGCACUAAACUUAUUAAUAA (((((((...-----))))))).(..(------(((((((.....))))))))..).......((((((.(((..((((((((......)))).))))..)))))))))........... ( -35.60) >DroSim_CAF1 19981 114 - 1 UACAUAUGUGUGUAUGUAUGUAGUUAU------AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAGCCGUGGCUUUGGCAUGUGUAGACACUAAACUUAUUAAUAA (((((((((....))))))))).(..(------(((((((.....))))))))..).......((((((..((.(((((((((......)))).)))))))..))))))........... ( -31.00) >DroEre_CAF1 18728 104 - 1 --GGUAU--------GUAUGUAGUGAUAUUCGUAGUCCAUCAAAUGUGGGCUACGAAUCUUU-UUUAGUCGCUCG-----CCGAAGCUUUGGCAUGUGUAGACACUAAACUUAUUAAUAA --((((.--------((...(((((..(((((((((((((.....)))))))))))))....-......(((..(-----((((....)))))..)))....))))).)).))))..... ( -31.50) >DroYak_CAF1 18322 112 - 1 UACAUAU--------GUAUGUAGUAAUACGCGCAGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAGCCGGAGCUUUGGCAUGUGUAGACACUAAACUUAUUAAUAA .......--------(..((((....))))..)(((((((.....)))))))...........((((((..((.((((((((((....))))).)))))))..))))))........... ( -28.40) >consensus UACAUAU________GUAUGUAGUUAU______AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAGCCGUGGCUUUGGCAUGUGUAGACACUAAACUUAUUAAUAA (((((((........)))))))...........(((((((.....)))))))...........((((((..((.(((((((((......)))).)))))))..))))))........... (-21.74 = -23.26 + 1.52)

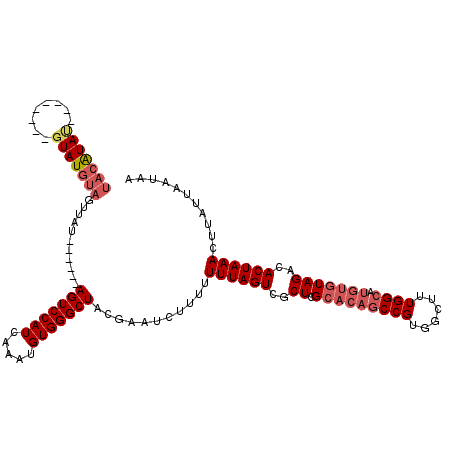

| Location | 19,703,787 – 19,703,882 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -16.45 |
| Energy contribution | -18.83 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19703787 95 - 23771897 GAG-----------CUGGCCGAGCUGAUAUAUACGUAGUAUACAUAUU--------UAUGUAGUUAU------AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAG ...-----------(((..(((((.(((...(((((((.........)--------)))))).(..(------(((((((.....))))))))..)...........))))))))..))) ( -25.70) >DroSec_CAF1 17864 109 - 1 GAGCUGGCAGAGCGCUGGCCGAGCUGAUAUAUACGUAGUAUACAUACGUA-----GUAUGUAGUUAU------AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAG ...(((((.(((((((((...((((.(((((((((((.......))))).-----)))))))))).(------(((((((.....))))))))...........)))).))))))).))) ( -33.90) >DroSim_CAF1 20021 107 - 1 -------CAGAGCGCUGGCCGAGCUGAUAUAUACGUAGUAUACAUAUGUGUGUAUGUAUGUAGUUAU------AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAG -------.......(((..(((((...((((((((((.(((((....))))))))))))))).(..(------(((((((.....))))))))..)..............)))))..))) ( -33.20) >DroYak_CAF1 18362 87 - 1 GAGC--------------------UGCUAUAUAC-----AUACAUAU--------GUAUGUAGUAAUACGCGCAGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAG ((((--------------------((((((((((-----((....))--------)))))))))).....((.(((((((.....))))))).))...............))))...... ( -28.80) >consensus GAG___________CUGGCCGAGCUGAUAUAUACGUAGUAUACAUAUG_______GUAUGUAGUUAU______AGUCCAUCAAAUGUGGGCUACGAAUCUUUUUUUAGUCGCUCGCACAG ..............(((..(((((.(((............(((((((........)))))))...........(((((((.....)))))))...............))))))))..))) (-16.45 = -18.83 + 2.37)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:29 2006