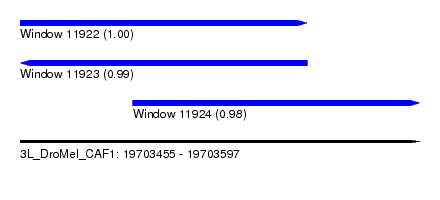
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,703,455 – 19,703,597 |
| Length | 142 |
| Max. P | 0.999614 |

| Location | 19,703,455 – 19,703,557 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.35 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19703455 102 + 23771897 AUAACAUUUGUUGUUGUACGCCUUGCCAUAUAUACAUACAUACA--UAUAUGUGCGUGAUUGUUGUUCUUUUUAAGCAAACAAAA-AAAA-----------AGAG--CAACAA--AACAG ((((((.....)))))).......(((((((((..........)--)))))).))((..((((((((((((((............-))))-----------))))--))))))--.)).. ( -24.60) >DroSec_CAF1 17530 117 + 1 AUAACAUUUGUUGUUGUACGCCUUGCCAUAUAUACAUACAUAUGUACAUAUAUGCGUAAUUGUUGUUCUUUUUCAGCAAACCAAA-AAAAAAAAAACAACAAGAG--CAACAAAAAACAG ......(((((((((.......(((((((((((((((....)))))..)))))).))))((((((((.(((((............-..))))).)))))))).))--)))))))...... ( -24.74) >DroSim_CAF1 19698 106 + 1 AUAACAUUUGUUGUUGUACGCCUUGCCAUAUAUACAUACAUAUGUACAUAUGUGCGUGAUUGUUGUUCUUUUUAAGCAAACCAAA-AAAA-----------AGAG--UAACAAAAAACAG ((((((.....)))))).......(((((((.(((((....))))).))))).))((..((((((((((((((............-))))-----------))))--))))))...)).. ( -23.30) >DroEre_CAF1 18480 88 + 1 AUAACAUUUGUUGUUGUACGCC-UGCCAUAUAUACG--CGUACA--UACAUAUGCGUGAUUGUUGCUCUU-UUAAGCAAACAG------------------------CUACG--AAACAG ....(((.(((...(((((((.-............)--))))))--.))).)))((((.((((((((...-...))).)))))------------------------.))))--...... ( -21.52) >DroYak_CAF1 18050 102 + 1 AUAACAUUUGUUGUUGUACGCCUUGCCAUAUAU--------------ACAUAUGCGUGAUUGUUGUUCUUUUUAAGCAAACAGCAACAAA--ACAGCAACAAAACAGCCACC--AAACAG ......(((((((((((.....(..((((((..--------------..))))).)..)((((((((.(((......))).)))))))).--))))))))))).........--...... ( -23.60) >consensus AUAACAUUUGUUGUUGUACGCCUUGCCAUAUAUACAUACAUACA__CAUAUAUGCGUGAUUGUUGUUCUUUUUAAGCAAACAAAA_AAAA___________AGAG__CAACAA_AAACAG ((((((.....)))))).....(((((((((((..............))))))).))))(((((..........)))))......................................... ( -8.14 = -8.34 + 0.20)

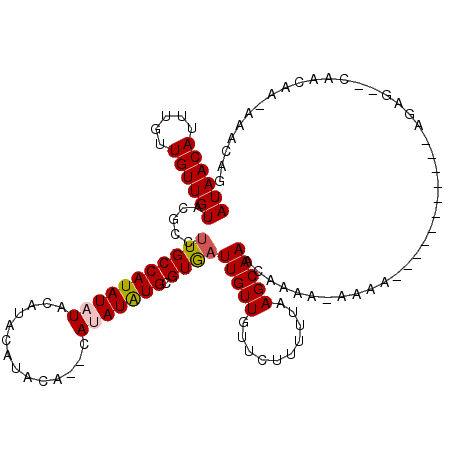
| Location | 19,703,455 – 19,703,557 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -8.23 |
| Energy contribution | -9.14 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19703455 102 - 23771897 CUGUU--UUGUUG--CUCU-----------UUUU-UUUUGUUUGCUUAAAAAGAACAACAAUCACGCACAUAUA--UGUAUGUAUGUAUAUAUGGCAAGGCGUACAACAACAAAUGUUAU .((((--((((((--.(((-----------((((-....(....)..))))))).))))))..((((...((((--(((((....))))))))).....))))..))))........... ( -22.10) >DroSec_CAF1 17530 117 - 1 CUGUUUUUUGUUG--CUCUUGUUGUUUUUUUUUU-UUUGGUUUGCUGAAAAAGAACAACAAUUACGCAUAUAUGUACAUAUGUAUGUAUAUAUGGCAAGGCGUACAACAACAAAUGUUAU ..((((.((((((--...(((((((((((((((.-...........))))))))))))))).(((((.(((((((((((....))))))))))).....))))))))))).))))..... ( -34.30) >DroSim_CAF1 19698 106 - 1 CUGUUUUUUGUUA--CUCU-----------UUUU-UUUGGUUUGCUUAAAAAGAACAACAAUCACGCACAUAUGUACAUAUGUAUGUAUAUAUGGCAAGGCGUACAACAACAAAUGUUAU .((((..(((((.--.(((-----------((((-...((....)).)))))))..)))))..((((.(((((((((((....))))))))))).....))))..))))........... ( -25.10) >DroEre_CAF1 18480 88 - 1 CUGUUU--CGUAG------------------------CUGUUUGCUUAA-AAGAGCAACAAUCACGCAUAUGUA--UGUACG--CGUAUAUAUGGCA-GGCGUACAACAACAAAUGUUAU .((((.--(((..------------------------((((((((((..-..))))))........((((((((--((....--)))))))))))))-))))...))))........... ( -21.70) >DroYak_CAF1 18050 102 - 1 CUGUUU--GGUGGCUGUUUUGUUGCUGU--UUUGUUGCUGUUUGCUUAAAAAGAACAACAAUCACGCAUAUGU--------------AUAUAUGGCAAGGCGUACAACAACAAAUGUUAU ......--.(((((...(((((((.(((--.((((((.(.(((......))).).))))))..((((...(((--------------.......)))..))))))).))))))).))))) ( -22.30) >consensus CUGUUU_UUGUUG__CUCU___________UUUU_UUUUGUUUGCUUAAAAAGAACAACAAUCACGCAUAUAUA__CAUAUGUAUGUAUAUAUGGCAAGGCGUACAACAACAAAUGUUAU ..((((.((((((..........................((((.........)))).......((((.((((((..(((....)))..)))))).....)))).)))))).))))..... ( -8.23 = -9.14 + 0.91)

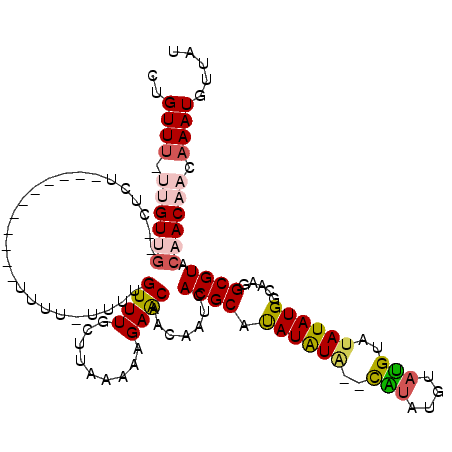
| Location | 19,703,495 – 19,703,597 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -14.92 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19703495 102 + 23771897 UACA--UAUAUGUGCGUGAUUGUUGUUCUUUUUAAGCAAACAAAA-AAAA-----------AGAG--CAACAA--AACAGCAGCGAUUAAUGGCUGAGCGAUAUGCAAACAGCAAAUGGC ..((--(.....(((((..((((((((((((((............-))))-----------))))--))))))--.))..((((........)))).)))...(((.....))).))).. ( -25.30) >DroSec_CAF1 17570 117 + 1 UAUGUACAUAUAUGCGUAAUUGUUGUUCUUUUUCAGCAAACCAAA-AAAAAAAAAACAACAAGAG--CAACAAAAAACAGCAGCGAUUAAUGGCUGAGCGAUAUGCAAACAGCAAAUGGC ((((((......)))))).(((((((((((...............-..............)))))--)))))).......((((........)))).((.((.(((.....))).)).)) ( -22.65) >DroSim_CAF1 19738 106 + 1 UAUGUACAUAUGUGCGUGAUUGUUGUUCUUUUUAAGCAAACCAAA-AAAA-----------AGAG--UAACAAAAAACAGCAGCGAUUAAUGGCUGAGCGAUAUGCAAACAGCAAAUGGC (((((((....))))))).((((((((((((((............-))))-----------))))--)))))).......((((........)))).((.((.(((.....))).)).)) ( -25.20) >DroEre_CAF1 18517 91 + 1 UACA--UACAUAUGCGUGAUUGUUGCUCUU-UUAAGCAAACAG------------------------CUACG--AAACAGCAGCGAUUAAUGGCAGAGCGAUAUGCAAACAGCAAAUGGC ....--..(((.(((.(((((((((((.((-((.(((.....)------------------------))..)--))).)))))))))))...(((........))).....))).))).. ( -25.20) >DroYak_CAF1 18083 109 + 1 -------ACAUAUGCGUGAUUGUUGUUCUUUUUAAGCAAACAGCAACAAA--ACAGCAACAAAACAGCCACC--AAACAGCAGCGAUUAAUGGCAGAGCGAUAUGCAAACAGCAAAUGGC -------.(((.(((((..((((((((.((((...((.....))...)))--).)))))))).)).((((..--((.(......).))..))))...((.....)).....))).))).. ( -21.20) >consensus UACA__CAUAUAUGCGUGAUUGUUGUUCUUUUUAAGCAAACAAAA_AAAA___________AGAG__CAACAA_AAACAGCAGCGAUUAAUGGCUGAGCGAUAUGCAAACAGCAAAUGGC .............((.(((((((((((...................................................)))))))))))...))...((.((.(((.....))).)).)) (-14.92 = -14.60 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:27 2006