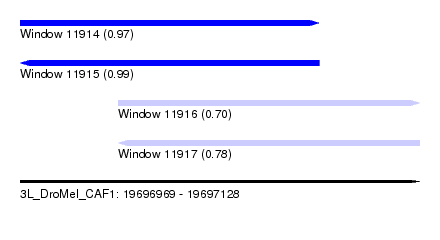
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,696,969 – 19,697,128 |
| Length | 159 |
| Max. P | 0.986181 |

| Location | 19,696,969 – 19,697,088 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -16.04 |
| Energy contribution | -15.84 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19696969 119 + 23771897 UGCACGUGUGUAUACACACAUACAUUCAUAC-UUAUGUAUGUGCGUACAAAUUGUCGAGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAU .(((((((((....)))))((((((......-..))))))))))......(((((((.(((((((.................................))))...))).))))))).... ( -26.11) >DroSec_CAF1 11355 116 + 1 UGCACGUGUGUAUACACACACACACACACACAUUU----CAUACAUACAAAUUGUCGGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAU .....((((((......))))))............----.................((((((((............(((....(((....)))....)))..(......))))))))).. ( -19.50) >DroSim_CAF1 13293 113 + 1 UGCACGUGUGUAUACACACACACACACAC---UUU----CGUACAUACAAAUUGUCGGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAU .....((((((......))))))......---...----.(.(((.......))))((((((((............(((....(((....)))....)))..(......))))))))).. ( -19.60) >DroEre_CAF1 11850 101 + 1 UGCACGUGUGUAUACACAUACU-------------------GACGUACAAAUUGUCUGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAU .((..((((((....)))))).-------------------............((((((((.((((........................))))))))))))))................ ( -20.26) >DroYak_CAF1 11957 101 + 1 UGCACGUGUGUACACACAUACU-------------------AACAUACAAAUUGUCUGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAU .((..((((((....)))))).-------------------............((((((((.((((........................))))))))))))))................ ( -20.26) >consensus UGCACGUGUGUAUACACACACACA__CA____UU_______UACAUACAAAUUGUCGGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAU .((..((((((....))))))................................(((.((((.((((........................)))))))).)))))................ (-16.04 = -15.84 + -0.20)

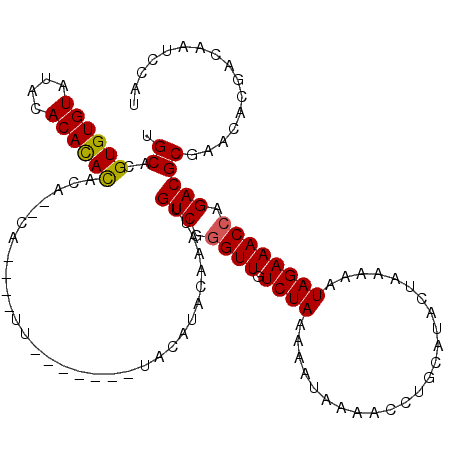
| Location | 19,696,969 – 19,697,088 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19696969 119 - 23771897 AUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACUCGACAAUUUGUACGCACAUACAUAA-GUAUGAAUGUAUGUGUGUAUACACACGUGCA (..((((((((.(((.((((((((.........)))).))))..((((.......))))))).))))))))..).....(((((....-)))))..((((((((((....)))))))))) ( -36.90) >DroSec_CAF1 11355 116 - 1 AUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCCGACAAUUUGUAUGUAUG----AAAUGUGUGUGUGUGUGUGUGUAUACACACGUGCA (..((((((((.(((.((((((((.........)))).))))..((((.......))))))).))))))))..).......----...((..((((((((((....))))))))))..)) ( -34.60) >DroSim_CAF1 13293 113 - 1 AUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCCGACAAUUUGUAUGUACG----AAA---GUGUGUGUGUGUGUGUAUACACACGUGCA (..((((((((.(((.((((((((.........)))).))))..((((.......))))))).))))))))..)(..(((.----...---)))..)(..((((((....))))))..). ( -35.60) >DroEre_CAF1 11850 101 - 1 AUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCAGACAAUUUGUACGUC-------------------AGUAUGUGUAUACACACGUGCA ......((.((((.....((((((((((((.....((((.........))))..)))).))))))))......)))).)-------------------)((((((((....)))))))). ( -26.90) >DroYak_CAF1 11957 101 - 1 AUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCAGACAAUUUGUAUGUU-------------------AGUAUGUGUGUACACACGUGCA (..(((((((......((((((((.........)))).)))).(((((((....))))..))).)))))))..).....-------------------.((((((((....)))))))). ( -27.70) >consensus AUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCCGACAAUUUGUAUGUA_______AA____UG__UGUGUGUGUGUAUACACACGUGCA (..((((((((.(((.((((((((.........)))).))))..((((.......))))))).))))))))..).........................((((((((....)))))))). (-23.52 = -23.88 + 0.36)

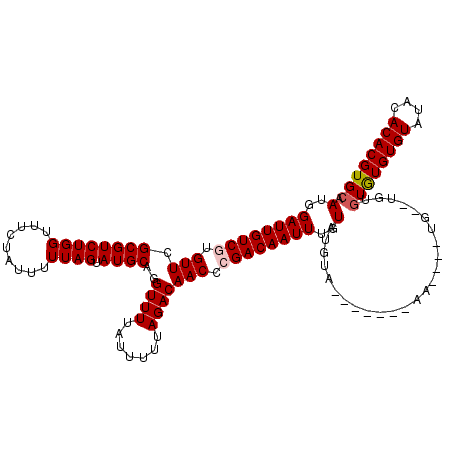
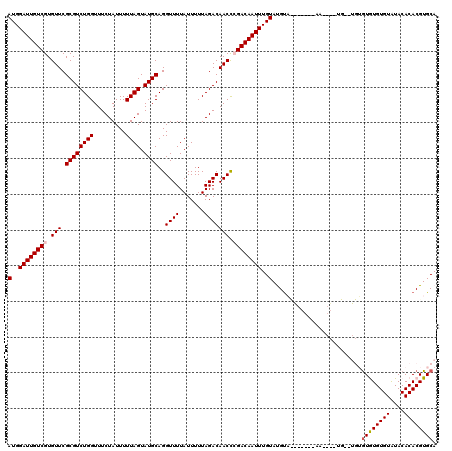
| Location | 19,697,008 – 19,697,128 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.41 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -18.12 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19697008 120 + 23771897 GUGCGUACAAAUUGUCGAGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAUUGGCCGUUGGAUAAACCAACACGACAUUACAUUGCACAUA (((((.......(((((.((((((............(((....(((....)))....)))..(......))))))).........(((((.....))))).)))))......)))))... ( -24.52) >DroSec_CAF1 11391 120 + 1 AUACAUACAAAUUGUCGGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAUUGGCCGUUGGAUAAACCAACACGACAUUACAUUGCACAUA ............((((((((((((............(((....(((....)))....)))..(......))))))))).......(((((.....)))))..)))).............. ( -22.20) >DroSim_CAF1 13326 120 + 1 GUACAUACAAAUUGUCGGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAUUGGCCGUUGGAUAAACCAACACGACAUUACAUUGCACAUA ((.((.......((((((((((((............(((....(((....)))....)))..(......))))))))).......(((((.....)))))..))))......)).))... ( -23.52) >DroEre_CAF1 11872 119 + 1 -GACGUACAAAUUGUCUGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAUUGGCCGUUGGAUAAACCAACACGACAUUACGUUGCACAUG -((((((......((((((((.((((........................)))))))))))).......((.(((....)))...(((((.....))))).))....))))))....... ( -24.36) >DroYak_CAF1 11979 119 + 1 -AACAUACAAAUUGUCUGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAUUGGCCGUUGGAUAAACCAACACGACAUUACAUUGCACAUA -............((((((((.((((........................))))))))))))((((......(((....)))...(((((.....)))))...........))))..... ( -22.26) >consensus _UACAUACAAAUUGUCGGGUUGUCUAAAAAUAAAACCUGCAUACUAAAAAUAGAAACCAGACGCGAACACGACAAUCCAUUGGCCGUUGGAUAAACCAACACGACAUUACAUUGCACAUA ............((((((((((((............(((....(((....)))....)))..(......))))))))).......(((((.....)))))..)))).............. (-18.12 = -18.72 + 0.60)

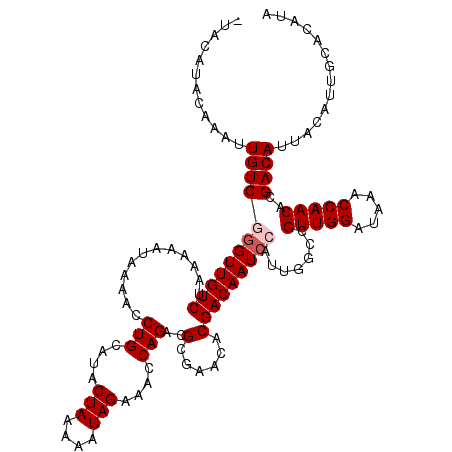
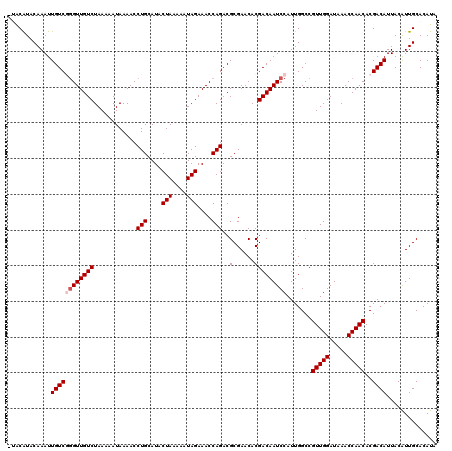
| Location | 19,697,008 – 19,697,128 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.41 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.26 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19697008 120 - 23771897 UAUGUGCAAUGUAAUGUCGUGUUGGUUUAUCCAACGGCCAAUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACUCGACAAUUUGUACGCAC ...((((........(((((..(((.....))))))))..(..((((((((.(((.((((((((.........)))).))))..((((.......))))))).))))))))..)..)))) ( -31.90) >DroSec_CAF1 11391 120 - 1 UAUGUGCAAUGUAAUGUCGUGUUGGUUUAUCCAACGGCCAAUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCCGACAAUUUGUAUGUAU ...(((((.......(((((..(((.....))))))))..(..((((((((.(((.((((((((.........)))).))))..((((.......))))))).))))))))..).))))) ( -28.70) >DroSim_CAF1 13326 120 - 1 UAUGUGCAAUGUAAUGUCGUGUUGGUUUAUCCAACGGCCAAUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCCGACAAUUUGUAUGUAC ...(((((.......(((((..(((.....))))))))..(..((((((((.(((.((((((((.........)))).))))..((((.......))))))).))))))))..).))))) ( -29.80) >DroEre_CAF1 11872 119 - 1 CAUGUGCAACGUAAUGUCGUGUUGGUUUAUCCAACGGCCAAUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCAGACAAUUUGUACGUC- .((((((((((....(((.((((((((........)))))))))))...)))......((((((((((((.....((((.........))))..)))).))))))))....))))))).- ( -30.90) >DroYak_CAF1 11979 119 - 1 UAUGUGCAAUGUAAUGUCGUGUUGGUUUAUCCAACGGCCAAUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCAGACAAUUUGUAUGUU- ((((.(((((.((.((.(.((((((.....))))))).)).)).))))))))).....((((((((((((.....((((.........))))..)))).))))))))............- ( -28.10) >consensus UAUGUGCAAUGUAAUGUCGUGUUGGUUUAUCCAACGGCCAAUGGAUUGUCGUGUUCGCGUCUGGUUUCUAUUUUUAGUAUGCAGGUUUUAUUUUUAGACAACCCGACAAUUUGUAUGUA_ .((((((((.....((((.((((((.....))))))......((.(((((((....)).((((....(((....)))....))))...........))))))).))))..)))))))).. (-26.46 = -26.26 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:19 2006