| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
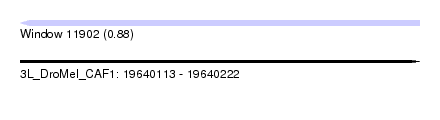
| Location | 19,640,113 – 19,640,222 |
| Length | 109 |
| Max. P | 0.875265 |

| Location | 19,640,113 – 19,640,222 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
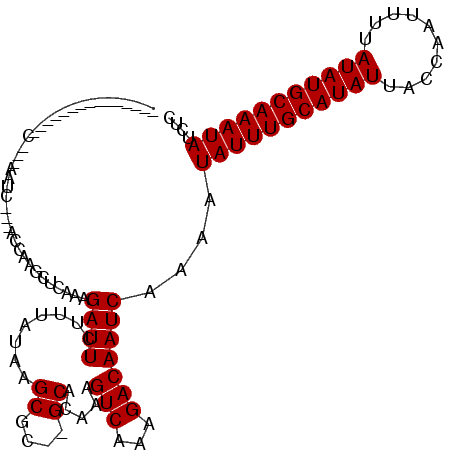
| Structure conservation index | 0.74 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
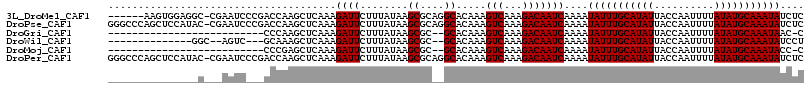
>3L_DroMel_CAF1 19640113 109 - 23771897 ------AAGUGGAGGC-CGAAUCCCGACCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUC ------..(((..((.-((.....)).))..(((.((((...))))...))).....)))...(((...))).......((((((((((((..........))))))))))))... ( -19.10) >DroPse_CAF1 56913 115 - 1 GGGCCCAGCUCCAUAC-CGAAUCCCGACCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUC ..(((..(((..(((.-.(((((..((......))...)))))..))).)))...))).....(((...))).......((((((((((((..........))))))))))))... ( -23.20) >DroGri_CAF1 60542 87 - 1 --------------------------CCCAAGCUCAAAGAUUCUUUAUAAGCGC--GCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAAC-C --------------------------.....(((.((((...))))...)))..--.......(((...)))........(((((((((((..........)))))))))))..-. ( -12.50) >DroWil_CAF1 82019 95 - 1 --------------GGC--AGUC---GCAAAGCUCAAAGAUUCUUUAUAAGCGC--GCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCCU --------------(((--.(((---((...(((.((((...))))...)))))--).))...))).............((((((((((((..........))))))))))))... ( -16.60) >DroMoj_CAF1 60966 87 - 1 --------------------------CCCGAGCUCAAAGAUUCUUUAUAAGCGC--GCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUACC-C --------------------------..((.(((.((((...))))...))).)--)......(((...)))........(((((((((((..........)))))))))))..-. ( -13.50) >DroPer_CAF1 56791 115 - 1 GGGCCCAGCUCCAUAC-CGAAUCCCGACCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUC ..(((..(((..(((.-.(((((..((......))...)))))..))).)))...))).....(((...))).......((((((((((((..........))))))))))))... ( -23.20) >consensus _______________C___AAUC___ACCAAGCUCAAAGAUUCUUUAUAAGCGC__GCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUC ......................................((((........((....)).....(((...)))))))....(((((((((((..........))))))))))).... (-13.37 = -13.37 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:04 2006