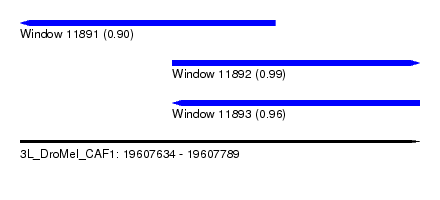
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,607,634 – 19,607,789 |
| Length | 155 |
| Max. P | 0.990497 |

| Location | 19,607,634 – 19,607,733 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.08 |
| Mean single sequence MFE | -47.86 |
| Consensus MFE | -34.32 |
| Energy contribution | -36.00 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19607634 99 - 23771897 ACUCAGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUCCGCAUCUGGAGCUGCUG------------CUUCGGAUACCGAUUCACUUGCUUCUUCGCCUGACGAAGGAG .(((.(.(((((.(((.((((((((..((((((......))))(((((((((....)------------))))))))..))..)).)))))).))))))))..)....))) ( -44.00) >DroSec_CAF1 22645 105 - 1 ACUGCGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUCCGCUUCUGGAGCUGCUG------CUGCGGCUUCGGAUACUGAUUCACUUGCUUCUUCGCCCGACGAAGCAG .((((...(((....))((((((((...(((((.((((((.(..(((((((((((..------..)))))))))))..).)))))).)))))))))))))...)...)))) ( -51.80) >DroSim_CAF1 22490 105 - 1 ACUCCGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUCCGCUUCUGGAGCUGCUG------CUGCGGCUUCGGAUACUGAUUCACUUGCUUCUUCGCCCGACGAAGCAG .((.((..(((....))((((((((...(((((.((((((.(..(((((((((((..------..)))))))))))..).)))))).))))))))))))))..)).))... ( -49.30) >DroEre_CAF1 22844 111 - 1 ACCCCGGAAACGGAAGUGCCGAGGAUCCGGCGGCUGGAUCCGCAUCUGGAGCUGCUGCUGCUGCUGCGGCUUCGGACCCCGAUUCACCUGCUUCUACGCCCGAGGAAGCAG ...(((....)))....((((......))))((.((((((.(..(((((((((((.((....)).)))))))))))..).))))))))(((((((.(....).))))))). ( -52.60) >DroYak_CAF1 22387 105 - 1 ACUCCGGAAACGGAAGUGCCGAGGACGCGGCGGCUGGAUCUGCACCUGGACCUGAUG------CUGCAGAUUCGGAUCUUGAUUCACCUGCUGCUACGCCCGAGGAAGCAG ..((((....)))).((.((..((.((..(((((.(((((((((.(..........)------.)))))))))((((....))))....)))))..)).))..))..)).. ( -41.60) >consensus ACUCCGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUCCGCAUCUGGAGCUGCUG______CUGCGGCUUCGGAUACUGAUUCACUUGCUUCUUCGCCCGACGAAGCAG ....(((..((....)).(((((((...(((((.((((((.(..(((((((((((..........)))))))))))..).)))))).)))))))))))))))......... (-34.32 = -36.00 + 1.68)

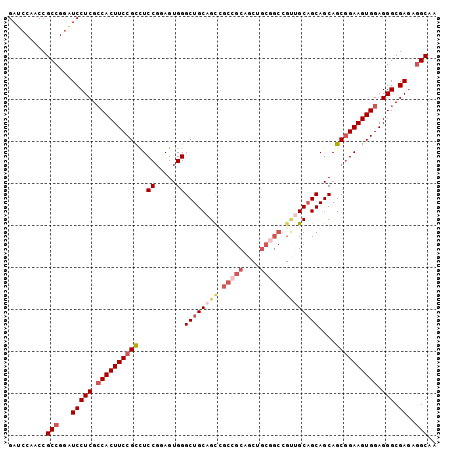
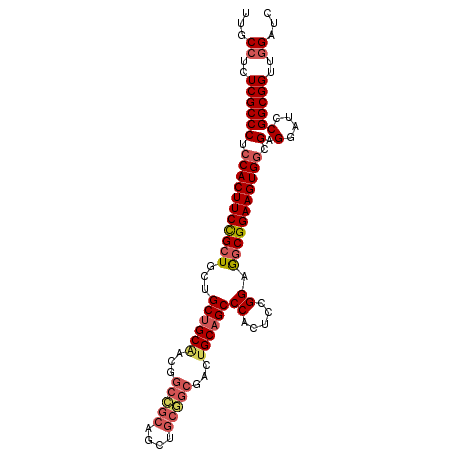
| Location | 19,607,693 – 19,607,789 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -53.52 |
| Consensus MFE | -40.34 |
| Energy contribution | -43.40 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19607693 96 + 23771897 GAUCCAACCGCCGGAUCCUCGCCACUUCCGCCUCCUGAGUGGGCUGCAGCUGCCGCCGC------CGCAGCAGCAGCAGCAGAAGUGGAGGGCGAGACGCAA (((((.......)))))((((((.((.((((..((.....))(((((.(((((.((...------.)).))))).)))))....))))))))))))...... ( -46.40) >DroSec_CAF1 22710 102 + 1 GAUCCAACCGCCGGAUCCUCGCCACUUCCGCCUCCGCAGUGGGCUGCAGUCGCCGCAGCUGCGGCCGUUGCAGCAGCAGCGGAAGUGGAGGGCGAGAGGCAA (((((.......)))))((((((.((.((((.(((((.((..(((((((..(((((....)))))..))))))).)).))))).))))))))))))...... ( -60.20) >DroSim_CAF1 22555 102 + 1 GAUCCAACCGCCGGAUCCUCGCCACUUCCGCCUCCGGAGUGGGCUGCAGUCGCCGCAGCUGCGGCCGUUGCAGCAGCAGCGGAAGUGGAGGGCGAGAGGCAA (((((.......)))))((((((.((.((((.((((..((..(((((((..(((((....)))))..))))))).))..)))).))))))))))))...... ( -56.90) >DroYak_CAF1 22452 96 + 1 GAUCCAGCCGCCGCGUCCUCGGCACUUCCGUUUCCGGAGUGGGCUGCAGCCGCAGC---UGCCGC---CGCCGCAGCAGCGGAAGUGGAGGGCGACAGGCAA .........(((.(((((((..((((((((((..(((....(((.(((((....))---))).))---).)))....)))))))))))))))))...))).. ( -50.60) >consensus GAUCCAACCGCCGGAUCCUCGCCACUUCCGCCUCCGGAGUGGGCUGCAGCCGCCGCAGCUGCGGCCGUUGCAGCAGCAGCGGAAGUGGAGGGCGAGAGGCAA .........(((...(((((.((((((((((..((.....))((((((((.(((((....))))).))))))))....)))))))))).))).))..))).. (-40.34 = -43.40 + 3.06)

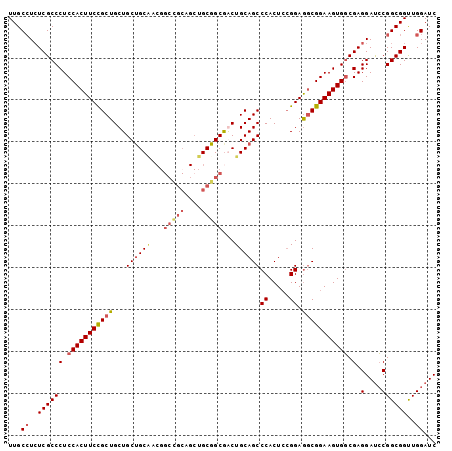
| Location | 19,607,693 – 19,607,789 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -52.90 |
| Consensus MFE | -38.92 |
| Energy contribution | -40.67 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19607693 96 - 23771897 UUGCGUCUCGCCCUCCACUUCUGCUGCUGCUGCUGCG------GCGGCGGCAGCUGCAGCCCACUCAGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUC ......((((((((((.((((((((((.((((((((.------...)))))))).))))).......))))).)).)).))))))((((((.....)))))) ( -50.51) >DroSec_CAF1 22710 102 - 1 UUGCCUCUCGCCCUCCACUUCCGCUGCUGCUGCAACGGCCGCAGCUGCGGCGACUGCAGCCCACUGCGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUC ......((((((((((.(((((((....((((((...(((((....)))))...)))))).....))))))).)).)).))))))((((((.....)))))) ( -55.00) >DroSim_CAF1 22555 102 - 1 UUGCCUCUCGCCCUCCACUUCCGCUGCUGCUGCAACGGCCGCAGCUGCGGCGACUGCAGCCCACUCCGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUC ...((..((((((.(((((((((((...((((((...(((((....)))))...))))))((.....)).))))))))))).).(....))))))..))... ( -52.00) >DroYak_CAF1 22452 96 - 1 UUGCCUGUCGCCCUCCACUUCCGCUGCUGCGGCG---GCGGCA---GCUGCGGCUGCAGCCCACUCCGGAAACGGAAGUGCCGAGGACGCGGCGGCUGGAUC ..(((.(((((((((((((((((((((.((.(((---((....---))))).)).))))).......(....))))))))..))))..))))))))...... ( -54.10) >consensus UUGCCUCUCGCCCUCCACUUCCGCUGCUGCUGCAACGGCCGCAGCUGCGGCGACUGCAGCCCACUCCGGAGGCGGAAGUGGCGAGGAUCCGGCGGUUGGAUC ...((..((((((.(((((((((((...((((((...(((((....)))))...))))))((.....)).))))))))))).).(....))))))..))... (-38.92 = -40.67 + 1.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:55 2006