
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,605,690 – 19,605,783 |
| Length | 93 |
| Max. P | 0.869862 |

| Location | 19,605,690 – 19,605,783 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -14.22 |
| Energy contribution | -13.75 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19605690 93 + 23771897 GCGGAAUGGGUAAGGAGUU-----AGGUGUUGUCAUUAAUUAUUAAAGUGUUCUUGUAUUUUUAGUUCUGGCUCAGUCCAGCAACUCCUCUCAUUCCG .(((((((((..(((((((-----.((....((((..(((((..((((((......))))))))))).)))).....))...)))))))))))))))) ( -26.70) >DroSec_CAF1 20675 85 + 1 GCGGAGUGAGUGAGGAGUU-----AGUUGUUGUCAUUAAUUAUUAAAG--------UUUUCUUCGUUAUGGCUCAGUCCAGCGACUCCUCUCUUUCCG .((((..(((.((((((((-----.((((..(((((............--------...........)))))......)))))))))))).))))))) ( -23.80) >DroSim_CAF1 20477 93 + 1 GCGGAGUGAGUGAGGAGUU-----AGUUGUUGUCAUUAAUUAUUAAAGUGUUCUUGUUUUCUUCGUUAUGGCUUAGUCCAGCGACUCCUCUCUUUCCG .((((..(((.((((((((-----.((((..(((((.........(((....)))............)))))......)))))))))))).))))))) ( -24.90) >DroEre_CAF1 20447 93 + 1 GCGGAAUGAGUAAGGAGUU-----AGGUGUUGUAAUUAAUUAUUAAAGCGUUGUUGUAUUUUUAGUUAUUGCUCAGUCCAGCCACUCCUUUCUUUCCG .((((..(((.(((((((.-----.(((...(((((.(((((..((.(((....))).))..))))))))))........)))))))))).))))))) ( -23.30) >DroYak_CAF1 20417 93 + 1 GCGGAAUGAGAAAGGAGUU-----AGGUGUUGUAAUGAAUUAUAAAAGUGUUGUUGUAUUUUUAGUUAAGGCUCAGUCCAGCAACUCCUCUCUUUCCG .(((((.((((..(((((.-----...(((((...(((.(((((((((((......))))))))..)))...)))...)))))))))))))).))))) ( -24.70) >DroAna_CAF1 20729 96 + 1 GAGGAGCGGGUGGAAUGUCGCGGGUGGGGUUGUGUUUUAUUUACAAAC--UAGUUGGAUAUUUGGUUCUUGCUAAGUCCGGCGACGCCUUUGGUGACU ((((.(((..(.(((((((.(((.(((..(((((.......))))).)--)).)))))))))).)..)(((((......))))).))))))(....). ( -26.90) >consensus GCGGAAUGAGUAAGGAGUU_____AGGUGUUGUCAUUAAUUAUUAAAGUGUUGUUGUAUUUUUAGUUAUGGCUCAGUCCAGCGACUCCUCUCUUUCCG .(((((.(((..(((((((.........((((....))))..............................(((......))))))))))))).))))) (-14.22 = -13.75 + -0.47)

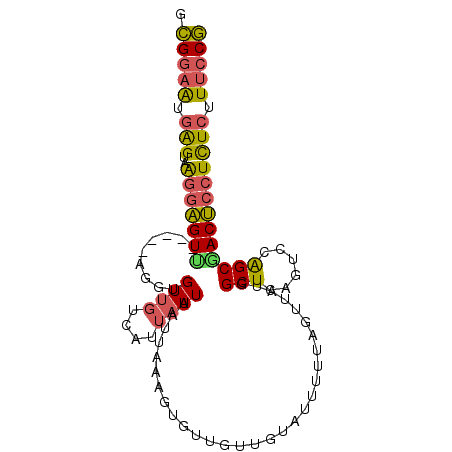

| Location | 19,605,690 – 19,605,783 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -18.58 |
| Consensus MFE | -8.20 |
| Energy contribution | -8.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19605690 93 - 23771897 CGGAAUGAGAGGAGUUGCUGGACUGAGCCAGAACUAAAAAUACAAGAACACUUUAAUAAUUAAUGACAACACCU-----AACUCCUUACCCAUUCCGC (((((((.(((((((((((((......))))............(((....)))....................)-----))))))))...))))))). ( -23.10) >DroSec_CAF1 20675 85 - 1 CGGAAAGAGAGGAGUCGCUGGACUGAGCCAUAACGAAGAAAA--------CUUUAAUAAUUAAUGACAACAACU-----AACUCCUCACUCACUCCGC ((((.((.(((((((.(((......)))......((((....--------))))....................-----.))))))).))...)))). ( -18.10) >DroSim_CAF1 20477 93 - 1 CGGAAAGAGAGGAGUCGCUGGACUAAGCCAUAACGAAGAAAACAAGAACACUUUAAUAAUUAAUGACAACAACU-----AACUCCUCACUCACUCCGC ((((.((.(((((((.(((......)))......((((............))))....................-----.))))))).))...)))). ( -17.70) >DroEre_CAF1 20447 93 - 1 CGGAAAGAAAGGAGUGGCUGGACUGAGCAAUAACUAAAAAUACAACAACGCUUUAAUAAUUAAUUACAACACCU-----AACUCCUUACUCAUUCCGC (((((.(((((((((....((...((((.....................))))((((.....)))).....)).-----.)))))))..)).))))). ( -17.50) >DroYak_CAF1 20417 93 - 1 CGGAAAGAGAGGAGUUGCUGGACUGAGCCUUAACUAAAAAUACAACAACACUUUUAUAAUUCAUUACAACACCU-----AACUCCUUUCUCAUUCCGC (((((((((((((((((((......)))......(((((............)))))..................-----)))))))))))..))))). ( -20.40) >DroAna_CAF1 20729 96 - 1 AGUCACCAAAGGCGUCGCCGGACUUAGCAAGAACCAAAUAUCCAACUA--GUUUGUAAAUAAAACACAACCCCACCCGCGACAUUCCACCCGCUCCUC .(((......)))(((((.((...(((...((........))...)))--((((.......)))).........)).)))))................ ( -14.70) >consensus CGGAAAGAGAGGAGUCGCUGGACUGAGCCAUAACUAAAAAUACAACAACACUUUAAUAAUUAAUGACAACACCU_____AACUCCUCACUCACUCCGC ((((.((.(((((((.(((......))).......................((....)).....................))))))).))...)))). ( -8.20 = -8.87 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:50 2006