| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,570,202 – 19,570,309 |
| Length | 107 |
| Max. P | 0.997988 |

| Location | 19,570,202 – 19,570,309 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19570202 107 + 23771897 CCGCCAUCAAAGACACACACUUUGUGUAGCACUAAAAGGAUAAAGGAUUCAA-------------ACACUGGUGCCUGCAAAAUGAAAGCGGCAUUCACCAUAAGAUCCUGCGAUGGCGG ((((((((....(((((.....))))).((.((....))....((((((...-------------.....((((((.((.........))))))))........)))))))))))))))) ( -34.29) >DroSec_CAF1 13656 107 + 1 CCGCCAUCAAGGACACGCACUUUGUGUAGCACUAAAAGGAUAAAGGAUUAAA-------------ACACUGGUGCCUACAAAAUGAAAGUGGCACUCACCAUGAGAUCCUGCGAUGGCGG ((((((((.(((((((.((.((((((..((((((..................-------------....)))))).)))))).))...)))...(((.....))).))))..)))))))) ( -36.15) >DroSim_CAF1 14648 107 + 1 CCGCCAUCAAGGACACACACUUUGUGUAGCACUAAAAGGAUAAAGGAUUCAA-------------ACACUGGUGCCUGCGAAAUGAAAGUGGCACUCACCAUGAGAUCCUGCGAUGGCGG ((((((((.((((....((((((.((((((((((....((........))..-------------....))))).))))).....))))))...(((.....))).))))..)))))))) ( -36.90) >DroEre_CAF1 16096 119 + 1 CCGCCAUCAAGGACACACACUUUGUGUAGCACUAACGGGAAAAAGGAUUCAAUUAAAGGCAGCUGUCACUGUUGCCUAA-AAACGCGAGCCGCACUCACCAUAAGAUCCUGUGAUGGCGG ((((((((.((..(((((.....)))).)..)).((((((....((..........(((((((.......)))))))..-....(((...))).....))......)))))))))))))) ( -40.20) >DroYak_CAF1 14032 118 + 1 CCGCCAUCAAGGACACACACUUUGUGUAGCACUAAA-AAAAGAAGGAUUUUAUUAAAAGCAGCUAUAACUGUUGCCUAG-AAAAGCGAACCGCACUCACCAUGAGAUCCUGCGAUGGCGG ((((((((....(((((.....))))).((.((...-...)).((((((((((.....(((((.......)))))....-....(((...))).......)))))))))))))))))))) ( -38.10) >consensus CCGCCAUCAAGGACACACACUUUGUGUAGCACUAAAAGGAUAAAGGAUUCAA_____________ACACUGGUGCCUAC_AAAUGAAAGCGGCACUCACCAUGAGAUCCUGCGAUGGCGG ((((((((....(((((.....)))))................(((((........................((((..............))))(((.....))))))))..)))))))) (-25.36 = -25.84 + 0.48)

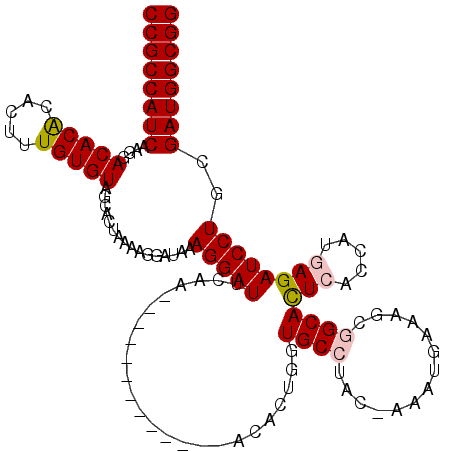

| Location | 19,570,202 – 19,570,309 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -27.92 |
| Energy contribution | -27.56 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19570202 107 - 23771897 CCGCCAUCGCAGGAUCUUAUGGUGAAUGCCGCUUUCAUUUUGCAGGCACCAGUGU-------------UUGAAUCCUUUAUCCUUUUAGUGCUACACAAAGUGUGUGUCUUUGAUGGCGG ((((((((((((((((...(((((..(((............)))..)))))....-------------..).)))).(((......))))))(((((.....))))).....)))))))) ( -33.60) >DroSec_CAF1 13656 107 - 1 CCGCCAUCGCAGGAUCUCAUGGUGAGUGCCACUUUCAUUUUGUAGGCACCAGUGU-------------UUUAAUCCUUUAUCCUUUUAGUGCUACACAAAGUGCGUGUCCUUGAUGGCGG (((((((((.(((((..(.((((....))))....((((((((.(((((.((...-------------..(((....))).....)).)))))..)))))))).).)))))))))))))) ( -36.20) >DroSim_CAF1 14648 107 - 1 CCGCCAUCGCAGGAUCUCAUGGUGAGUGCCACUUUCAUUUCGCAGGCACCAGUGU-------------UUGAAUCCUUUAUCCUUUUAGUGCUACACAAAGUGUGUGUCCUUGAUGGCGG (((((((((.((((((.(((..((.(((.((((.........((((((....)))-------------)))................)))).))).))..))).).)))))))))))))) ( -35.61) >DroEre_CAF1 16096 119 - 1 CCGCCAUCACAGGAUCUUAUGGUGAGUGCGGCUCGCGUUU-UUAGGCAACAGUGACAGCUGCCUUUAAUUGAAUCCUUUUUCCCGUUAGUGCUACACAAAGUGUGUGUCCUUGAUGGCGG (((((((((.(((((..(((.(((((..((((..(((((.-...(....)...))).)).)))..((((.(((......)))..)))))..)).)))...)))...)))))))))))))) ( -40.50) >DroYak_CAF1 14032 118 - 1 CCGCCAUCGCAGGAUCUCAUGGUGAGUGCGGUUCGCUUUU-CUAGGCAACAGUUAUAGCUGCUUUUAAUAAAAUCCUUCUUUU-UUUAGUGCUACACAAAGUGUGUGUCCUUGAUGGCGG (((((((((.((((((.(((.(((((..(.....((((..-..))))..((((....))))......................-....)..)).)))...))).).)))))))))))))) ( -39.50) >consensus CCGCCAUCGCAGGAUCUCAUGGUGAGUGCCGCUUUCAUUU_GUAGGCACCAGUGU_____________UUGAAUCCUUUAUCCUUUUAGUGCUACACAAAGUGUGUGUCCUUGAUGGCGG (((((((((.((((((.(((..((.(((.((((..........(((............................)))..........)))).))).))..))).).)))))))))))))) (-27.92 = -27.56 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:39 2006