| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,542,127 – 19,542,226 |
| Length | 99 |
| Max. P | 0.980227 |

| Location | 19,542,127 – 19,542,226 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.13 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -13.13 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
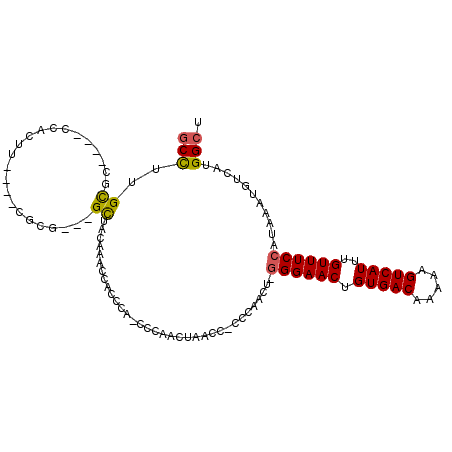
>3L_DroMel_CAF1 19542127 99 + 23771897 GCCUUGCGC----CCACUU----CGCG---GCGACAAAGCACCCA-CCCAACUAACCGCCCAACUUGGGAACUGUGACACAAAGUCAUUUGUUUCCAUAAAUGUCAUGGCU ((.((((((----((....----.).)---))).))).)).....-...........(((..((.(((((((.(((((.....)))))..))))))).....))...))). ( -28.50) >DroPse_CAF1 3585 87 + 1 -CUGUGCUC----CCACAU----CUCAUCGGUCUCCG-----CCCCCCCAG--------UG-AGG-GGGAACUGUGACAAAAAGUCAUUUGUUUCCAUGAAUGUCAGGGCA -...(((((----..((((----.((((.((......-----..(((((..--------..-.))-)))(((.(((((.....)))))..))).))))))))))..))))) ( -28.30) >DroSec_CAF1 13186 98 + 1 GCCUUGCGC----CCACUU----CGCG---GCUACAAGCCACCCA-CCCAACUAACCACCCAACU-GGGAACUGUGACAAAAAGUCAUUUGUUUCCAUAAAUGUCAUGGCU (((..(((.----......----)))(---((.....))).....-..................(-((((((.(((((.....)))))..)))))))..........))). ( -24.50) >DroEre_CAF1 4107 98 + 1 GCCGUGUGC----CCACUU----GGUG---GCUACCAACCACCCA-CCCAACUAACCGCCCAGCU-GGGAACUGUGACAAAAAGUCAUCUGUUUCCAUAAAUGUCAUGGCU ((((((.((----......----((((---(.......)))))..-..................(-((((((.(((((.....)))))..))))))).....)))))))). ( -29.50) >DroYak_CAF1 29834 98 + 1 GCCUUGCGU----CCACUU----CUUG---GCUACAAACCACGCA-CCCAACUAACCACCCAGCU-GGGAACUGUGACAAAAAGUCAUCUGUUUCCAUAAAUGUCAGGGCU (((((((((----......----..((---(.......)))....-..................(-((((((.(((((.....)))))..)))))))...))).)))))). ( -24.60) >DroAna_CAF1 5298 94 + 1 GCCUUGCAAGAUACCACACCCCCAUUG---GCCACCG-----CCA-CCCAA-------GCCAUCU-UGGAACUGUGACAAAAAGUCAUUUGUUUCCAUAAAUGUCAUGGCA (((...........((((.......((---((....)-----)))-.((((-------(....))-)))...)))).......(.(((((((....))))))).)..))). ( -21.80) >consensus GCCUUGCGC____CCACUU____CGCG___GCUACAAACCACCCA_CCCAACUAACC_CCCAACU_GGGAACUGUGACAAAAAGUCAUUUGUUUCCAUAAAUGUCAUGGCU (((..((.......................))..................................((((((.(((((.....)))))..))))))...........))). (-13.13 = -13.05 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:34 2006