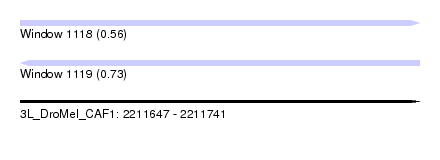
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,211,647 – 2,211,741 |
| Length | 94 |
| Max. P | 0.729617 |

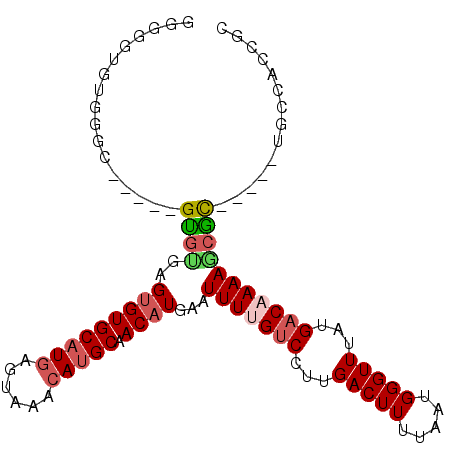
| Location | 2,211,647 – 2,211,741 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.13 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2211647 94 + 23771897 GCGGUGCCA-----GCGCUUUUGUCAUAAACCCAUUAAAAGUCAAGGACAAAAUUCAUGUUGCAUGUUUACUCAUGCACACUCACAC-----GCCCACACCACC ..((((...-----(((.(((((((....((.........))....)))))))....(((.(((((......))))))))......)-----))...))))... ( -21.90) >DroGri_CAF1 255795 90 + 1 ----GGGCA-----ACGUUUUUGUCAUAAACCCAUUAAAAGUCAAGGGCGAAAUUCAUGUUGCAUGUUUACUCAUGCACGCUUUUGC-----ACACAAGCGCAC ----(.((.-----..((((.......)))).........((((((((((..........((((((......)))))))))))))).-----))....)).).. ( -20.30) >DroSim_CAF1 263782 94 + 1 GCGGUGCCA-----GCGCUUUUGUCAUAAACCCAUUAAAAGUCAAGGACAAAAUUCAUGUUGCAUGUUUACUCAUGCACACUCACAC-----GCCCACACCACC ..((((...-----(((.(((((((....((.........))....)))))))....(((.(((((......))))))))......)-----))...))))... ( -21.90) >DroEre_CAF1 270185 94 + 1 GUGGUGGCA-----GCGCUUUUGUCAUAAACCCAUUAAAAGUCAAGGACAAAAUUCAUGUUGCAUGUUUACUCAUGCACACUCACAC-----ACCCACGCACUC (((((((..-----....(((((((....((.........))....)))))))....(((.(((((......)))))))).......-----..)))).))).. ( -20.90) >DroMoj_CAF1 275168 86 + 1 ----UAGCA-----ACGUUUUUGUCAUAAACCCAUUAAAAGUCAAGGACGAAAUUCAUGUUGCAUGUUUACUUAAGCACACCCACAC-----ACACAUAC---- ----..(((-----(((((((((((....((.........))....)))))))...))))))).(((((....))))).........-----........---- ( -14.40) >DroAna_CAF1 257338 104 + 1 GCAGCAGGAGGGGCAUGCUUUUGUCAUAAACCCAUUAAAAGUCAAGGACAAAAUUCAUGUUGCAUGUUUACUCAUGCACACUCGCACACUCCGCCCACACCUAC .....(((.(((((..(((((((............)))))))...(((.........(((.(((((......)))))))).........))))))).).))).. ( -25.37) >consensus GCGGUGGCA_____ACGCUUUUGUCAUAAACCCAUUAAAAGUCAAGGACAAAAUUCAUGUUGCAUGUUUACUCAUGCACACUCACAC_____ACCCACACCAAC ..................(((((((....((.........))....)))))))....(((.(((((......))))))))........................ (-11.60 = -11.13 + -0.47)

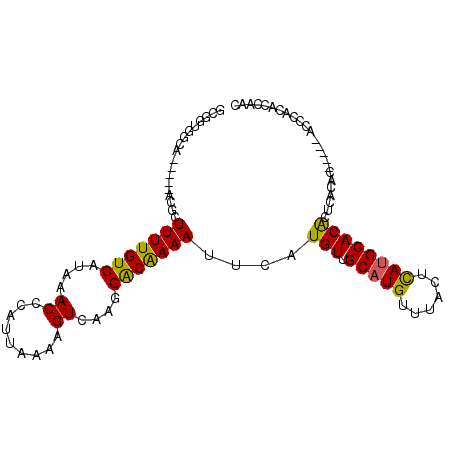
| Location | 2,211,647 – 2,211,741 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -15.23 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2211647 94 - 23771897 GGUGGUGUGGGC-----GUGUGAGUGUGCAUGAGUAAACAUGCAACAUGAAUUUUGUCCUUGACUUUUAAUGGGUUUAUGACAAAAGCGC-----UGGCACCGC .(((((((.(((-----((....(((((((((......))))).))))...(((((((...(((((.....)))))...)))))))))))-----).))))))) ( -38.90) >DroGri_CAF1 255795 90 - 1 GUGCGCUUGUGU-----GCAAAAGCGUGCAUGAGUAAACAUGCAACAUGAAUUUCGCCCUUGACUUUUAAUGGGUUUAUGACAAAAACGU-----UGCCC---- .(((((....))-----)))..((((((((((......)))))..(((((.....((((............)))))))))......))))-----)....---- ( -23.00) >DroSim_CAF1 263782 94 - 1 GGUGGUGUGGGC-----GUGUGAGUGUGCAUGAGUAAACAUGCAACAUGAAUUUUGUCCUUGACUUUUAAUGGGUUUAUGACAAAAGCGC-----UGGCACCGC .(((((((.(((-----((....(((((((((......))))).))))...(((((((...(((((.....)))))...)))))))))))-----).))))))) ( -38.90) >DroEre_CAF1 270185 94 - 1 GAGUGCGUGGGU-----GUGUGAGUGUGCAUGAGUAAACAUGCAACAUGAAUUUUGUCCUUGACUUUUAAUGGGUUUAUGACAAAAGCGC-----UGCCACCAC ..(((.((((..-----((((..(((((((((......))))).))))...(((((((...(((((.....)))))...)))))))))))-----..))))))) ( -30.90) >DroMoj_CAF1 275168 86 - 1 ----GUAUGUGU-----GUGUGGGUGUGCUUAAGUAAACAUGCAACAUGAAUUUCGUCCUUGACUUUUAAUGGGUUUAUGACAAAAACGU-----UGCUA---- ----.(((((.(-----(((((....(((....)))..)))))))))))......(((...(((((.....)))))...)))........-----.....---- ( -15.80) >DroAna_CAF1 257338 104 - 1 GUAGGUGUGGGCGGAGUGUGCGAGUGUGCAUGAGUAAACAUGCAACAUGAAUUUUGUCCUUGACUUUUAAUGGGUUUAUGACAAAAGCAUGCCCCUCCUGCUGC (((((.(.((((.....((((..(((((((((......))))).))))...(((((((...(((((.....)))))...)))))))))))))))).)))))... ( -37.80) >consensus GGGGGUGUGGGC_____GUGUGAGUGUGCAUGAGUAAACAUGCAACAUGAAUUUUGUCCUUGACUUUUAAUGGGUUUAUGACAAAAGCGC_____UGCCACCGC .................((((..(((((((((......))))).))))...(((((((...(((((.....)))))...))))))))))).............. (-15.23 = -15.82 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:39 2006