
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,526,717 – 19,526,886 |
| Length | 169 |
| Max. P | 0.888475 |

| Location | 19,526,717 – 19,526,824 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
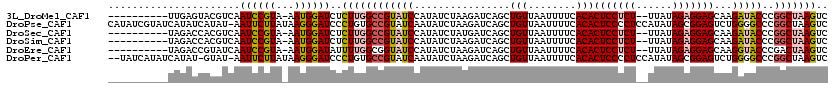
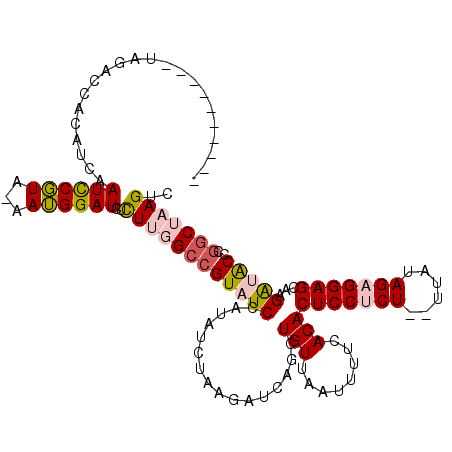
>3L_DroMel_CAF1 19526717 107 + 23771897 ----------UUGAGUACGUCAAUCCGUA-AAUGGAUCUCUUGGCCGUAUCCAUAUCUAAGAUCAGCUGUUAAUUUUCACACUCCUCU--UUAUAGAGGAGCAAGAUACCCGGCUAAGUC ----------............(((((..-..)))))..((((((((((((................(((........)))(((((((--....)))))))...)))))..))))))).. ( -26.30) >DroPse_CAF1 14540 119 + 1 CAUAUCGUAUCAUAUCAUAU-AAUUCUUAUAAGGGAUCCCUGUGCCGUAUCAAUAUCUAAGAUCAGCUGUUAAUUUUCACACUCCCCUCCAUAUAGCGGAGUCUGGGGCCCGGCUAAGUC ....................-....((((...(((.((((.(((....((.((((.((......)).)))).))...)))(((((.((......)).)))))..)))))))...)))).. ( -21.40) >DroSec_CAF1 14772 107 + 1 ----------UAGACCACGUCAAUCCGUA-AAUGGAUCUCUUGGCCGUAUCCAUAUCUAUGAUCAGCUGUUAAUUUUCACACUCCUCU--UUAUAGAGGAGCAAGAUACCCGGCUAAGUC ----------..(((...))).(((((..-..)))))..(((((((((((((((....)))....................(((((((--....)))))))...)))))..))))))).. ( -27.70) >DroSim_CAF1 18341 107 + 1 ----------UAGACCACGUCAAUCCGUA-AAUGGAUCUCUUGGCCGUAUCCAUAUCUAAGAUCAGCUGUUAAUUUUCACACUCCUCU--UUAUAGAGGAGCAAGAUACCCGGCUAAGUC ----------..(((...))).(((((..-..)))))..((((((((((((................(((........)))(((((((--....)))))))...)))))..))))))).. ( -27.10) >DroEre_CAF1 17575 107 + 1 ----------UAGACCGUAUCAAUCCGUA-AAUGGAUAUUUUGGCGGUAUCCAUAUCUAAGAUCAGCUGUUAAUUUUCACACUCCUCU--UUAUAGAGGAGCAAGGUACCCGACUAAGUC ----------......(((((.(((((..-..)))))...(((((((((((.........)))..))))))))........(((((((--....)))))))...)))))........... ( -23.80) >DroPer_CAF1 14596 116 + 1 --UAUCAUAUCAUAU-GUAU-AAUUCUUAUAAGGGAUCCCUGUGCCGUAUCAAUAUCUAAGAUCAGCUGUUAAUUUUCACACUCCCCUCCAUAUAGCGGAGUCUGGGGCCCGGCUAAGUC --.............-....-....((((...(((.((((.(((....((.((((.((......)).)))).))...)))(((((.((......)).)))))..)))))))...)))).. ( -21.40) >consensus __________UAGACCACAUCAAUCCGUA_AAUGGAUCUCUUGGCCGUAUCCAUAUCUAAGAUCAGCUGUUAAUUUUCACACUCCUCU__UUAUAGAGGAGCAAGAUACCCGGCUAAGUC ......................((((((...))))))..((((((((((((................(((........)))(((((((......)))))))...)))))..))))))).. (-19.83 = -20.22 + 0.39)
| Location | 19,526,717 – 19,526,824 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
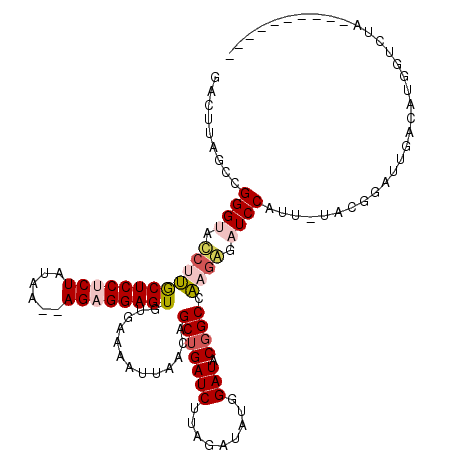
>3L_DroMel_CAF1 19526717 107 - 23771897 GACUUAGCCGGGUAUCUUGCUCCUCUAUAA--AGAGGAGUGUGAAAAUUAACAGCUGAUCUUAGAUAUGGAUACGGCCAAGAGAUCCAUU-UACGGAUUGACGUACUCAA---------- .........(((((((((((((((((....--)))))))(((........)))((((((((.......)))).)))))))))(((((...-...)))))....)))))..---------- ( -31.00) >DroPse_CAF1 14540 119 - 1 GACUUAGCCGGGCCCCAGACUCCGCUAUAUGGAGGGGAGUGUGAAAAUUAACAGCUGAUCUUAGAUAUUGAUACGGCACAGGGAUCCCUUAUAAGAAUU-AUAUGAUAUGAUACGAUAUG ..((((...((((((((.(((((.((......)).))))).))..........(((((((.........))).))))...)))..)))...)))).(((-(((...))))))........ ( -30.60) >DroSec_CAF1 14772 107 - 1 GACUUAGCCGGGUAUCUUGCUCCUCUAUAA--AGAGGAGUGUGAAAAUUAACAGCUGAUCAUAGAUAUGGAUACGGCCAAGAGAUCCAUU-UACGGAUUGACGUGGUCUA---------- ...........((((((.((((((((....--))))))))((((..............))))......))))))(((((...(((((...-...)))))....)))))..---------- ( -31.14) >DroSim_CAF1 18341 107 - 1 GACUUAGCCGGGUAUCUUGCUCCUCUAUAA--AGAGGAGUGUGAAAAUUAACAGCUGAUCUUAGAUAUGGAUACGGCCAAGAGAUCCAUU-UACGGAUUGACGUGGUCUA---------- (((......((((.((((((((((((....--)))))))(((........)))((((((((.......)))).))))))))).))))...-((((......)))))))..---------- ( -32.90) >DroEre_CAF1 17575 107 - 1 GACUUAGUCGGGUACCUUGCUCCUCUAUAA--AGAGGAGUGUGAAAAUUAACAGCUGAUCUUAGAUAUGGAUACCGCCAAAAUAUCCAUU-UACGGAUUGAUACGGUCUA---------- ((((..(((.(((.....((((((((....--))))))))((........)).)))(((((.....(((((((.........))))))).-...))))))))..))))..---------- ( -27.10) >DroPer_CAF1 14596 116 - 1 GACUUAGCCGGGCCCCAGACUCCGCUAUAUGGAGGGGAGUGUGAAAAUUAACAGCUGAUCUUAGAUAUUGAUACGGCACAGGGAUCCCUUAUAAGAAUU-AUAC-AUAUGAUAUGAUA-- ..((((...((((((((.(((((.((......)).))))).))..........(((((((.........))).))))...)))..)))...)))).(((-((((-....).)))))).-- ( -31.40) >consensus GACUUAGCCGGGUACCUUGCUCCUCUAUAA__AGAGGAGUGUGAAAAUUAACAGCUGAUCUUAGAUAUGGAUACGGCCAAGAGAUCCAUU_UACGGAUUGACAUGGUCUA__________ .........((((.((((((((((((......)))))))).............(((((((.........))).)))).)))).))))................................. (-21.33 = -21.83 + 0.50)

| Location | 19,526,786 – 19,526,886 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -27.67 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19526786 100 - 23771897 GCCCGCA-AAUGCCAAGAGAAACUUGAUCAAGAGUUAAGAGGUCGGCCGAAAGUUGUCCAUCUGACUUAGCCGGGUAUCUUGCUCCUCUAUAA--AGAGGAGU ((((((.-.....((((.....)))).....((((((.((((.(((.(....)))).)).)))))))).).))))).....((((((((....--)))))))) ( -33.70) >DroPse_CAF1 14619 103 - 1 GUCCGAAAAAUAUCAAGAGAAACUUGAUCAAGAGUUAAGAGGUCGGCAGAAAGUUUCGCAUCUGACUUAGCCGGGCCCCAGACUCCGCUAUAUGGAGGGGAGU (((((......((((((.....)))))).....((((((.((...((.((.....))))..))..))))))))))).....(((((.((......)).))))) ( -27.40) >DroSec_CAF1 14841 100 - 1 GCCCGCA-AAUGCCAAGAGAAACUUGAUCAAGAGUUAAGAGGUCGGCCGAAAGUUGCCCAUCUGACUUAGCCGGGUAUCUUGCUCCUCUAUAA--AGAGGAGU (((((..-.....((((.....)))).......((((((((((.((((....)..))).))))..))))))))))).....((((((((....--)))))))) ( -33.80) >DroSim_CAF1 18410 100 - 1 GCCCGCA-AAUGCCAAGAGAAACUUGAUCAAGAGUUAGGAGGUCGGCCGAAAGUUGCCCAUCUGACUUAGCCGGGUAUCUUGCUCCUCUAUAA--AGAGGAGU ((((((.-.....((((.....)))).....(((((((..((.(((.(....)))).))..))))))).).))))).....((((((((....--)))))))) ( -36.20) >DroEre_CAF1 17644 100 - 1 GCCCGCA-AAUAUCAAGAGAAACUUGAUCAAGAGUUAAGAGGUCGGCCGAAAGUUGUCCAUCUGACUUAGUCGGGUACCUUGCUCCUCUAUAA--AGAGGAGU ((((((.-...((((((.....))))))...((((((.((((.(((.(....)))).)).)))))))).).))))).....((((((((....--)))))))) ( -35.60) >DroPer_CAF1 14672 103 - 1 GUCCGAAAAAUAUCAAGAGAAACUUGAUCAAGAGUUAAGAGGUCGGCAGAAAGUUUCGCAUCUGACUUAGCCGGGCCCCAGACUCCGCUAUAUGGAGGGGAGU (((((......((((((.....)))))).....((((((.((...((.((.....))))..))..))))))))))).....(((((.((......)).))))) ( -27.40) >consensus GCCCGCA_AAUACCAAGAGAAACUUGAUCAAGAGUUAAGAGGUCGGCCGAAAGUUGCCCAUCUGACUUAGCCGGGUACCUUGCUCCUCUAUAA__AGAGGAGU (((((........((((.....)))).....((((((.((((.((((.....)))).)).))))))))...))))).....((((((((......)))))))) (-27.67 = -28.00 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:18 2006