| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,523,875 – 19,523,991 |
| Length | 116 |
| Max. P | 0.971023 |

| Location | 19,523,875 – 19,523,991 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.41 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -8.50 |
| Energy contribution | -9.34 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
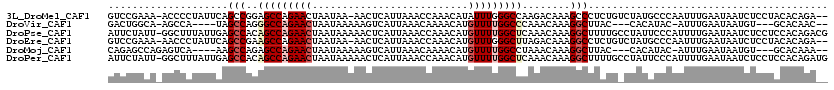
>3L_DroMel_CAF1 19523875 116 - 23771897 GUCCGAAA-ACCCCUAUUCAGCCGGAGCCAGAACUAAUAA-AACUCAUUAAACCAAACAUAUUUGGGCCAAGACAAAGCCCUCUGUCUAUGCCCAAUUUGAAUAAUCUCCUACACAGA-- ........-.....(((((((...(((.............-..)))................((((((..(((((........)))))..)))))).)))))))..............-- ( -19.06) >DroVir_CAF1 17519 106 - 1 GACUGGCA-AGCCA----UAGCCAGGGCCAGAACUAAUAAAAAGUCAUUAAACAAAACAUGUUUUGGCCCAAACAAAGGCUUAC---CACAUAC-AUUUGAAUAAUGU---GCACAAC-- ...(((.(-((((.----......((((((((((..........................)))))))))).......))))).)---))..(((-(((.....)))))---)......-- ( -28.91) >DroPse_CAF1 11773 119 - 1 AUUCUAUU-GGCUUUAUUGAGCCACAGCCAGAACUAAUAAAAACUCAUUAAACCAAACAUGUUUUGGCUCAAACAAAGGCUUUUGCCUAUUCCCAUUUUGAAUAAUCUCCUCCACAGACG ((((...(-(((((....)))))).(((((((((..........................))))))))).......((((....))))...........))))................. ( -24.37) >DroEre_CAF1 14563 116 - 1 GUCCGAAA-AACCCUAUUCAGCCGAAGCCAGAACUAAUAA-AACUCAUUAAACCAAACAUGUUUGGGCUUAGACAAAGGCCUCUGUCUAUGCCCAAUUUGAAUAAUCUCCUACACAGA-- ........-.....((((((((....))............-.....................((((((.((((((........)))))).))))))..))))))..............-- ( -21.10) >DroMoj_CAF1 17230 107 - 1 CAGAGCCAGAGUCA----AAGCCAGAGCCAGAACUAAUAAAAAGUCAUUAAACAAAACAUGUUUUGGCCUAAACAAAGGCUUAC---CACAUAC-AUUUGAAUAAUGU---GCACAAA-- ..(((((.(.((..----..)))((.((((((((..........................)))))))))).......)))))..---....(((-(((.....)))))---)......-- ( -19.57) >DroPer_CAF1 11834 119 - 1 AUUCUAUU-GGCUUUAUUGAGCCACAGCCAGAACUAAUAAAAACUCAUUAAACCAAACAUGUUUUGGCUCAAACAAAGGCUUUUGCCUAUUCCCAUUUUGAAUAAUCUCCUCCACAGAUG ((((...(-(((((....)))))).(((((((((..........................))))))))).......((((....))))...........)))).((((.......)))). ( -24.67) >consensus GUCCGAAA_AGCCAUAUUCAGCCAGAGCCAGAACUAAUAAAAACUCAUUAAACCAAACAUGUUUUGGCCCAAACAAAGGCUUAUG_CUAUACCCAAUUUGAAUAAUCUCCUACACAGA__ ....................(((..(((((((((..........................)))))))))........)))........................................ ( -8.50 = -9.34 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:15 2006