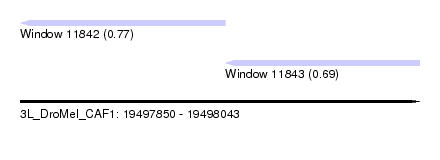
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,497,850 – 19,498,043 |
| Length | 193 |
| Max. P | 0.769201 |

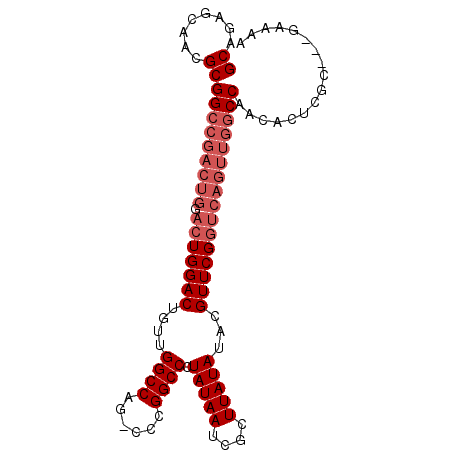
| Location | 19,497,850 – 19,497,949 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
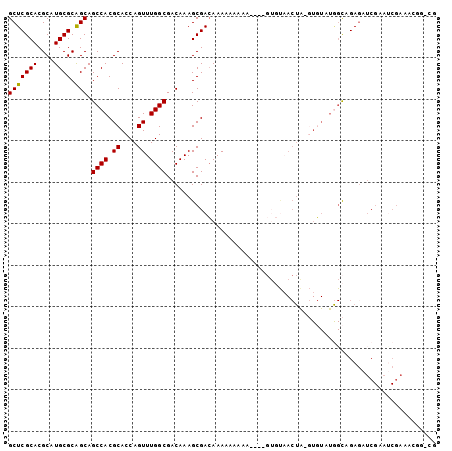
>3L_DroMel_CAF1 19497850 99 - 23771897 GCUCGCACGCAUGCGCAGCAGCCACGCACCAGUUUGGCGACAAAGCGACAAAAAAAAA----GUGUAACUAUGUGUUUGGCUGAGAUCGAAUCGAAGCGGUCG (((((((....)))).)))(((((.((((.((((..((......)).(((........----.)))))))..)))).)))))..(((((........))))). ( -28.10) >DroPse_CAF1 6692 80 - 1 GCUCGCACGCAUGCGCGGCAGCCACGCACCAGUUUGGCGACAAAGCGAAAAAAAAACA-------C-ACUAUAUUUAUAGCAGAAAAC--------------- ..((((.(((....)))(..((((.((....)).))))..)...))))..........-------.-.((((....))))........--------------- ( -17.30) >DroSec_CAF1 8510 103 - 1 GCUCGCACGCAUGCGCAGCAGCCACGCACCAGUUUGGCGACAAAGCGACAAAAAAAAAUAGAGUGUAACUAUGUGUUUGGCUGAGAUCGAAUCGAAACGGCCG (((((((....)))).))).((((.((....)).)))).(((.((..(((.............)))..)).)))....(((((...(((...)))..))))). ( -25.92) >DroSim_CAF1 8732 83 - 1 GCUCGCACGCAUGCGCAGCAGCCACGCACCAGUUUGGCGACAAAGCGACAAAAAAAAAAA--------------------UUGAGAUCGAAUCGAAACGGUCG (((((((....)))).))).((((.((....)).))))......................--------------------....(((((........))))). ( -20.70) >DroEre_CAF1 9012 95 - 1 GCUCGCACGCAUGCGCAGCAGCCACGCACCAGUUUGGCGACAAAGCGACAAAAAA-------GUGUAACUG-GUGUAUGGCAGAGAUCGAAUCGAAAAUUGAA (((((((....)))).))).((((..((((((((..((......)).(((.....-------.))))))))-)))..)))).....((((........)))). ( -32.40) >DroYak_CAF1 8709 88 - 1 GCUCGCACGCAUGCGCAGCAGCCACGCACCAGUUUGGCGACAAAGCGACAAAAAA-------GUGUAACUG-GUGUAUGGCAGAGAUCGAACCGAA------- (((((((....)))).))).((((..((((((((..((......)).(((.....-------.))))))))-)))..))))...............------- ( -32.00) >consensus GCUCGCACGCAUGCGCAGCAGCCACGCACCAGUUUGGCGACAAAGCGACAAAAAAAAA____GUGUAACUA_GUGUAUGGCAGAGAUCGAAUCGAAACGG_CG (((((((....)))).))).((((.((....)).))))................................................................. (-15.59 = -15.45 + -0.14)

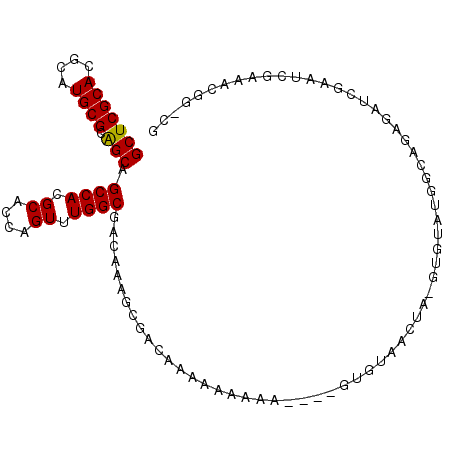
| Location | 19,497,949 – 19,498,043 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -29.45 |
| Energy contribution | -30.95 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19497949 94 - 23771897 GCAGAGCCACGCGGCCGACUGGACUGGACUGUUGGCCAG-CCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACAGUCGC---GAAAA ((.(.(((....))))..........(((((((((((((-((((..(.((((........)))).)..)))...))))))))))))))))---..... ( -38.70) >DroPse_CAF1 6772 97 - 1 GCAAGGCAGCGCGGCCGACUGGACUGGACUGUUGGCCAG-CCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACACUCGCACAGAAAA ((.((((...))(((((((((.(((((((....((((..-...)))).(((((....)))))...))))))))))))))))....)).))........ ( -36.20) >DroSim_CAF1 8815 94 - 1 GCAGAGCUACGCGGCCGACUGGACUGGACUGUUGGCCAG-CCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACAGUCGC---GAAAA ((.........(((((....)).)))(((((((((((((-((((..(.((((........)))).)..)))...))))))))))))))))---..... ( -38.10) >DroEre_CAF1 9107 93 - 1 GCAGAGCCACGCGGCCGACUGGACUGGACUGUUGGCCAG-CCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACAC-CGC---GAAAA ((...((...))(((((((((.(((((((....((((..-...)))).(((((....)))))...)))))))))))))))).....-.))---..... ( -35.60) >DroWil_CAF1 5735 75 - 1 GCA-----ACGCGG----------UGGACUGAUGGCCAGGCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAA--GUCGG---GU--- ...-----....((----------(.(((((((((((......)))).(((((....)))))........))))))).)))..--.....---..--- ( -22.10) >DroPer_CAF1 6605 97 - 1 GCAAGGCAGCGCGGCCGACUGGACUGGACUGUUGGCCAG-CCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACACUCGCACAGAAAA ((.((((...))(((((((((.(((((((....((((..-...)))).(((((....)))))...))))))))))))))))....)).))........ ( -36.20) >consensus GCAGAGCAACGCGGCCGACUGGACUGGACUGUUGGCCAG_CCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACACUCGC___GAAAA ((........))(((((((((.(((((((....((((......)))).(((((....)))))...))))))))))))))))................. (-29.45 = -30.95 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:07 2006