
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,497,335 – 19,497,451 |
| Length | 116 |
| Max. P | 0.935723 |

| Location | 19,497,335 – 19,497,430 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -16.87 |
| Consensus MFE | -11.85 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19497335 95 + 23771897 CAUUU--------------AUUUUUUCUGCAGAGCUACACGCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCU-UAAAAGCAUGCGGGUAUGCAAAAAAUAAA---- ..(((--------------((((((..(((...((.....)).(((.((((((((........))))..........(((-....)))..)))).)))))))))))))))---- ( -20.80) >DroVir_CAF1 6279 108 + 1 AAUUUUAAAUUUGGCUUGCAUUUUUUUGCCACAGCUACACGCAAUGGUCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAU------AUGCAAAAAAUAAAAAUA ..(((((..(((.((.(((........((((..((.....))..)))).((((((..................)))))).......))).------..)).)))..)))))... ( -16.87) >DroPse_CAF1 6128 92 + 1 CAUUU--------------AUUUUUU-UCCAGAGCUACACGCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAUACGAGUAUGCAAAC---AAA---- .....--------------.......-.(((..((.....))..)))..((((((..................)))))).......(((((....)))))....---...---- ( -15.07) >DroGri_CAF1 6106 96 + 1 AAUUUCAAAG----C-UACAUU--UUUGCCACAGCAACACGCAACGGUCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAU------AUGCAAAAUAAAA----- ..(((.((((----(-.((...--.((((....))))..(((.......)))......................)).))))).)))((..------..)).........----- ( -12.90) >DroAna_CAF1 5496 96 + 1 CAUUU--------------AUUUUUUUUCCAAAGCUACACGCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAUACAAGUAUGCCAGGAAAAAA---- .....--------------...(((((((((((((.((..((.......))((((........)))).......)).)))))....(((((....)))))..))))))))---- ( -20.50) >DroPer_CAF1 5971 92 + 1 CAUUU--------------AUUUUUU-UCCAGAGCUACACGCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAUACGAGUAUGCAAAC---AAA---- .....--------------.......-.(((..((.....))..)))..((((((..................)))))).......(((((....)))))....---...---- ( -15.07) >consensus CAUUU______________AUUUUUUUUCCAGAGCUACACGCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAUACGAGUAUGCAAAAAA_AAA____ ............................(((..((.....))..)))..((((((..................)))))).......(((((....))))).............. (-11.85 = -12.19 + 0.33)

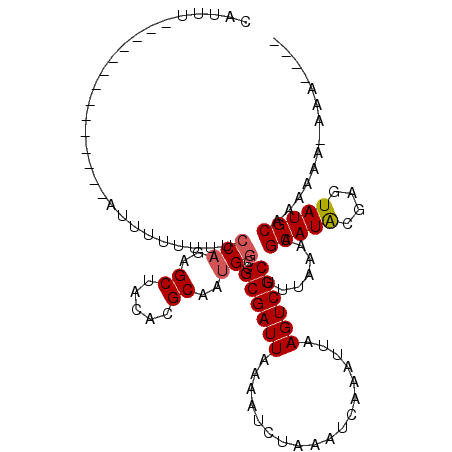
| Location | 19,497,361 – 19,497,451 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -13.62 |
| Consensus MFE | -9.75 |
| Energy contribution | -9.72 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
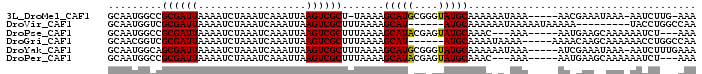
>3L_DroMel_CAF1 19497361 90 + 23771897 GCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCU-UAAAAGCAUGCGGGUAUGCAAAAAAUAAA-----AACGAAAUAAA-AAUCUUG-AAA (((.((.((((((((........))))..........(((-....)))..)))).)))))..........-----...........-.......-... ( -16.20) >DroVir_CAF1 6319 83 + 1 GCAAUGGUCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAU------AUGCAAAAAAUAAAAAUAAAAA---------UACCUGGCCAA ....(((((((((((..................)))))).......((..------..))..................---------.....))))). ( -13.67) >DroPse_CAF1 6153 87 + 1 GCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAUACGAGUAUGCAAAC---AAA-----AAUGAAGCAAAAAAUCU---AAA ((.......((((((..................)))))).......(((((....)))))....---...-----......)).........---... ( -13.17) >DroGri_CAF1 6139 87 + 1 GCAACGGUCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAU------AUGCAAAAUAAAA-----AAAACAAGCAAAAAACCUGGCCAA .....((((((((((..................)))))).......((..------..)).........-----..................)))).. ( -12.97) >DroYak_CAF1 8231 92 + 1 GCAAUGGCAGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAUGCGGGUAUGCAAAAAAUAAA-----AUCGAAAUAAA-AAUCUUUGAAA ((....))(((((((..................)))))))......(((((....)))))..........-----.(((((.....-....))))).. ( -12.57) >DroPer_CAF1 5996 87 + 1 GCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAUACGAGUAUGCAAAC---AAA-----AAUGAAGCAAAAAAUCU---AAA ((.......((((((..................)))))).......(((((....)))))....---...-----......)).........---... ( -13.17) >consensus GCAAUGGCCGCGAUUAAAAUCUAAAUCAAAUUAAGUCGCUUUAAAAGCAU_CG_GUAUGCAAAAAAUAAA_____AAAGAAGCAAA_AAUCU_G_AAA .........((((((..................)))))).......(((((....)))))...................................... ( -9.75 = -9.72 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:05 2006