
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,496,528 – 19,496,666 |
| Length | 138 |
| Max. P | 0.994673 |

| Location | 19,496,528 – 19,496,628 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.50 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -17.97 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
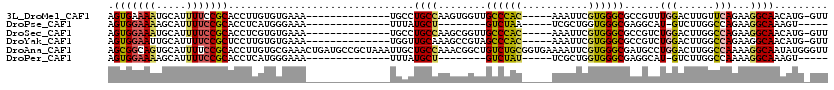
>3L_DroMel_CAF1 19496528 100 + 23771897 AGUGAAAAUGCAUUUUCCGCACCUUGUGUGAAA--------------UGCCUGCCAAGUGGUUGCCCAC-----AAAUUCGUGGGCGCCGUUUGGACUUGUUCAGAAGGCAACAUG-GUU .........((((((..(((.....)))..)))--------------)))..((((.((....((((((-----......))))))(((.((((((....)))))).))).)).))-)). ( -37.30) >DroPse_CAF1 5403 87 + 1 AGUGGAAAAGCAUUUUCCGCACCUCAUGGGAAA--------------UUUAUGCU--------GUCUAA-----UCGCUGGUGGGCGAGGCAU-GUCUUGGCCAGAAGGCAAAGU----- .(((((((.....))))))).(((..(((..((--------------..((((((--------......-----(((((....))))))))))-)..))..)))..)))......----- ( -26.70) >DroSec_CAF1 7208 100 + 1 AGUGGAAAUGCAUUUUCCGCACCUCGUGUGAAA--------------UGCCUGCCAAGCGGUUGCCCAC-----AAAUUCGUGGGCGCCGUCUGGACUUGGCCAGAAGGCAACAUG-GUU .(((((((.....)))))))...((((((....--------------(((((((((((((((.((((((-----......))))))))))......))))))....))))))))))-).. ( -43.40) >DroYak_CAF1 7370 100 + 1 AGUGGAAUUGCAUUUUCCGCUCCUUGUGUGAAA--------------UGGUUGCAAAGCCGUAGCCCAC-----AAAUUCGUGGGCGCCGUCUGGACUUGGCCAGAAGGCAACAUG-GUU (((((((.......)))))))((.(((.....(--------------(((((....)))))).((((((-----......))))))(((.(((((......))))).))).))).)-).. ( -41.30) >DroAna_CAF1 4733 120 + 1 AGCGGCAGUGCAUUUUCCGCACCUUGUGCGAAACUGAUGCCGCUAAAUUGCUGCCAAACGGCUGUCUGCGGUGAAAAUUCGUGGGCGAUGCCUGGACUUGGCCAAAAGGCAAUAUGGGUU (((((((...((.((((.((((...)))))))).)).)))))))..(((((((((((.((((((((..(((.......)))..))))..))).)...))))).....))))))....... ( -44.80) >DroPer_CAF1 5261 87 + 1 AGUGGAAAAGCAUUUUCCGCACCUCAUGGGAAA--------------UUUAUGCU--------GUCUAU-----UCGCUGGUGGGCGAGGCAU-GUCUUGGCCAAAAGGCAAAGU----- ((((((..((((((((((.((.....)))))))--------------...)))))--------.)))))-----).(((..(((.(((((...-.))))).)))...))).....----- ( -27.70) >consensus AGUGGAAAUGCAUUUUCCGCACCUCGUGUGAAA______________UGCCUGCCAAGCGGUUGCCCAC_____AAAUUCGUGGGCGACGCCUGGACUUGGCCAGAAGGCAACAUG_GUU .(((((((.....)))))))...............................((((........((((((...........))))))......(((......)))...))))......... (-17.97 = -17.83 + -0.14)
| Location | 19,496,561 – 19,496,666 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -43.14 |
| Consensus MFE | -36.41 |
| Energy contribution | -35.53 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
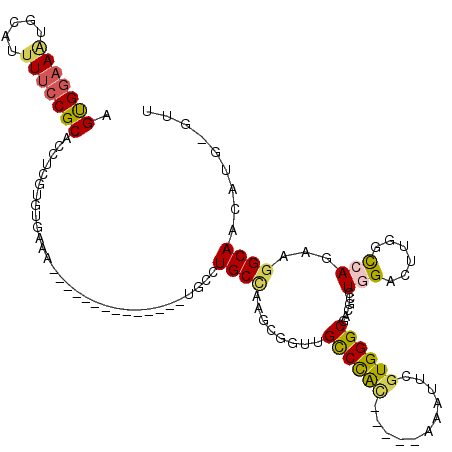
>3L_DroMel_CAF1 19496561 105 + 23771897 UGCCUGCCAAGUGGUUGCCCACAAAUUCGUGGGCGCCGUUUGGACUUGUUCAGAAGGCAACAUGGUUUAGUAACCGCCGCGCUUAAUGGCACAGUUAUGAUUUCC ((((......(((((.((((((......))))))(((.((((((....)))))).))).....((((....))))))))).......)))).............. ( -37.62) >DroSec_CAF1 7241 105 + 1 UGCCUGCCAAGCGGUUGCCCACAAAUUCGUGGGCGCCGUCUGGACUUGGCCAGAAGGCAACAUGGUUUAGUAACCGCUGCGCUUAAUGGCACAGUUAUGAUUUCU ((((.((((.((((((((((((......))))))(((.(((((......))))).))).............)))))))).)).....)))).............. ( -43.00) >DroYak_CAF1 7403 104 + 1 UGGUUGCAAAGCCGUAGCCCACAAAUUCGUGGGCGCCGUCUGGACUUGGCCAGAAGGCAACAUGGUUUAGUAGCCGCUGCGCU-AAUGGCACAGUUAUGAUUUCU .((((((.(((((((.((((((......))))))(((.(((((......))))).)))...))))))).))))))((((.((.-....)).)))).......... ( -48.80) >consensus UGCCUGCCAAGCGGUUGCCCACAAAUUCGUGGGCGCCGUCUGGACUUGGCCAGAAGGCAACAUGGUUUAGUAACCGCUGCGCUUAAUGGCACAGUUAUGAUUUCU ...(((....(((((.((((((......))))))(((.(((((......))))).))).....((((....)))))))))(((....))).)))........... (-36.41 = -35.53 + -0.88)


| Location | 19,496,561 – 19,496,666 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -36.47 |
| Consensus MFE | -32.15 |
| Energy contribution | -32.27 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
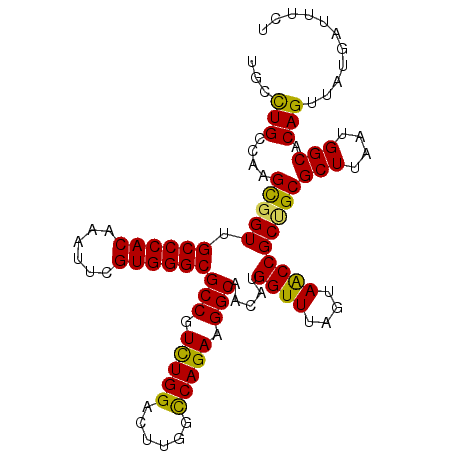
>3L_DroMel_CAF1 19496561 105 - 23771897 GGAAAUCAUAACUGUGCCAUUAAGCGCGGCGGUUACUAAACCAUGUUGCCUUCUGAACAAGUCCAAACGGCGCCCACGAAUUUGUGGGCAACCACUUGGCAGGCA ...........((((((......)))))).((((....))))....(((((.(..(....(((.....)))(((((((....)))))))......)..).))))) ( -33.00) >DroSec_CAF1 7241 105 - 1 AGAAAUCAUAACUGUGCCAUUAAGCGCAGCGGUUACUAAACCAUGUUGCCUUCUGGCCAAGUCCAGACGGCGCCCACGAAUUUGUGGGCAACCGCUUGGCAGGCA ..............(((((........(((((((.............(((.(((((......))))).)))(((((((....))))))))))))))))))).... ( -40.60) >DroYak_CAF1 7403 104 - 1 AGAAAUCAUAACUGUGCCAUU-AGCGCAGCGGCUACUAAACCAUGUUGCCUUCUGGCCAAGUCCAGACGGCGCCCACGAAUUUGUGGGCUACGGCUUUGCAACCA ...........((((((....-.)))))).((....((((((.....(((.(((((......))))).)))(((((((....)))))))...)).))))...)). ( -35.80) >consensus AGAAAUCAUAACUGUGCCAUUAAGCGCAGCGGUUACUAAACCAUGUUGCCUUCUGGCCAAGUCCAGACGGCGCCCACGAAUUUGUGGGCAACCGCUUGGCAGGCA ((((.......((((((......)))))).(((.((........)).))))))).(((((((((....)).(((((((....)))))))....)))))))..... (-32.15 = -32.27 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:04 2006