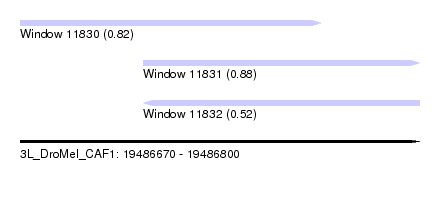
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,486,670 – 19,486,800 |
| Length | 130 |
| Max. P | 0.882833 |

| Location | 19,486,670 – 19,486,768 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.05 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19486670 98 + 23771897 UAAAAUUGCUGUGGGUGACGGUUAGGAAUUUAGUGGCCAUCGGAUAAGCAACAAUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUAAG-AAAUGCAAA ........(((..(((.((.............)).)))..)))....(((.(((((......))---)))....((..-.(((....)))..)-)..)))... ( -19.92) >DroPse_CAF1 85495 87 + 1 --AUAUU---UUAAGGAAUGGUUA------CAUUG-----CUGUUAAACAAUAAUAUUUGUUGACCUUUUUUUUUCGUUAAGCGAAAGUUAAGCAAACGAAAA --.....---..((((((((((..------....)-----))))).(((((......)))))..))))...((((((((.(((....))).....)))))))) ( -15.60) >DroSec_CAF1 126940 98 + 1 UUAAAAUGCAGUGGGUGAUGGUUAGGAAUUCAUUGGCCAUCGGCUAAGCAACAAUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUGAG-AUAUGCAAA ......(((..(((.((((((((((.......)))))))))).))).)))..............---(((.(.(((..-.(((....)))..)-)).).))). ( -27.30) >DroSim_CAF1 127075 98 + 1 UAAAAUUGCAGUGGGUGAUGGUUAGGAAUUUAUUGGCAAUCGGCUAAGCAACAAUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUGAG-AUGUGCAAA .....(((((..........((((((((((..(((((.....))))).....))))))))))..---......(((..-.(((....)))..)-)).))))). ( -22.40) >DroEre_CAF1 112322 97 + 1 -AAAAUGACUGCGGGUGAAGGCUAGCCAUUCAUUGGCCAUCGGCUGAGCCACGUUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUAGG-AAAUGCAAA -....(((..((.((((((((....)).)))))).))..)))((....((.((.((((..(...---)..)))).)).-.(((....))).))-....))... ( -26.40) >DroYak_CAF1 165254 93 + 1 -AAAAUGACAGUGGGUGAAGGCUAGGA----AUUGGCUAUCGGCUGAUCCAAGUUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUAAG-AAAUGCAAA -.......((((..(((((((((.(((----.(..((.....))..)))).)))))))..)).)---)))....((..-.(((....)))..)-)........ ( -23.60) >consensus _AAAAUUGCAGUGGGUGAUGGUUAGGAAUUCAUUGGCCAUCGGCUAAGCAACAAUUUUUGGCAA___UUGGAAAUCGA_GAGCGAAAGCUAAG_AAAUGCAAA ...........(((.(((.((((((.......)))))).))).)))..............(((.......(....)....(((....))).......)))... (-12.46 = -12.05 + -0.41)


| Location | 19,486,710 – 19,486,800 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -14.76 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19486710 90 + 23771897 CGGAUAAGCAACAAUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUAAG-AAAUGCAAACUUUCACU-UUGAGAGAAAGUCUCGAAAAAUAA (((((..(((.(((((......))---)))....((..-.(((....)))..)-)..)))...(((((...-..)))))...))).))........ ( -19.10) >DroSec_CAF1 126980 90 + 1 CGGCUAAGCAACAAUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUGAG-AUAUGCAAACUUUCUCU-UUGAGUGAAAGUCUCGAAAAAUAA ..((((((........))))))..---.......((((-((((....)))...-........((((((.(.-....).)))))))))))....... ( -21.30) >DroSim_CAF1 127115 90 + 1 CGGCUAAGCAACAAUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUGAG-AUGUGCAAACUUUCACU-UUGAGUGAAAGUCUCGAAAAAUAA ..((((((........))))))..---.......((((-((((....)))...-........((((((((.-....)))))))))))))....... ( -25.30) >DroEre_CAF1 112361 89 + 1 CGGCUGAGCCACGUUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUAGG-AAAUGCAAACUUUCACU-UUGAGUGAAAGUCU-AAAAAAUAA .(((...)))..(((((((((...---(((.(..((..-.(((....)))..)-)..).)))((((((((.-....))))))))))-))))))).. ( -24.60) >DroYak_CAF1 165289 89 + 1 CGGCUGAUCCAAGUUUUUUGGCAA---UUGGAAAUCGA-GAGCGAAAGCUAAG-AAAUGCAAACUUUCACU-UUAAGUGAAAGUCU-AAAAAAUAU .((.....))..(((((((((...---(((.(..((..-.(((....)))..)-)..).)))((((((((.-....))))))))))-))))))).. ( -22.90) >DroPer_CAF1 85605 95 + 1 CUGUUAAACAAUAAUAUUUGUUGACCUUUUUUUUUCGUUAAGCGAAAGUUAAGCAAACGAAAACUUUCACUUUUGAGUGAAAGUCGUG-CAAAUGU .............((((((((((((......((((((((.(((....))).....)))))))).(((((((....))))))))))).)-))))))) ( -22.80) >consensus CGGCUAAGCAACAAUUUUUGGCAA___UUGGAAAUCGA_GAGCGAAAGCUAAG_AAAUGCAAACUUUCACU_UUGAGUGAAAGUCUCGAAAAAUAA ..((((((........))))))..................(((....)))............(((((((((....)))))))))............ (-14.76 = -14.98 + 0.23)



| Location | 19,486,710 – 19,486,800 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19486710 90 - 23771897 UUAUUUUUCGAGACUUUCUCUCAA-AGUGAAAGUUUGCAUUU-CUUAGCUUUCGCUC-UCGAUUUCCAA---UUGCCAAAAAUUGUUGCUUAUCCG .......((((((...........-.(((((((((.......-...)))))))))))-))))....(((---((......)))))........... ( -15.90) >DroSec_CAF1 126980 90 - 1 UUAUUUUUCGAGACUUUCACUCAA-AGAGAAAGUUUGCAUAU-CUCAGCUUUCGCUC-UCGAUUUCCAA---UUGCCAAAAAUUGUUGCUUAGCCG .......((((((((((.....))-)).(((((((.......-...)))))))..))-))))....(((---((......)))))........... ( -14.30) >DroSim_CAF1 127115 90 - 1 UUAUUUUUCGAGACUUUCACUCAA-AGUGAAAGUUUGCACAU-CUCAGCUUUCGCUC-UCGAUUUCCAA---UUGCCAAAAAUUGUUGCUUAGCCG .......(((((((((((((....-.))))))))........-...(((....))))-))))....(((---((......)))))........... ( -18.20) >DroEre_CAF1 112361 89 - 1 UUAUUUUUU-AGACUUUCACUCAA-AGUGAAAGUUUGCAUUU-CCUAGCUUUCGCUC-UCGAUUUCCAA---UUGCCAAAAAACGUGGCUCAGCCG ........(-((((((((((....-.))))))))))).....-....(((.(((...-.))).......---..((((.......))))..))).. ( -18.20) >DroYak_CAF1 165289 89 - 1 AUAUUUUUU-AGACUUUCACUUAA-AGUGAAAGUUUGCAUUU-CUUAGCUUUCGCUC-UCGAUUUCCAA---UUGCCAAAAAACUUGGAUCAGCCG .........-((((((((((....-.))))))))))((....-...(((....))).-..((..(((((---.(........).))))))).)).. ( -17.20) >DroPer_CAF1 85605 95 - 1 ACAUUUG-CACGACUUUCACUCAAAAGUGAAAGUUUUCGUUUGCUUAACUUUCGCUUAACGAAAAAAAAAGGUCAACAAAUAUUAUUGUUUAACAG ..(((((-...((((((((((....))))))))))((((((.((.........))..)))))).............)))))............... ( -18.20) >consensus UUAUUUUUCGAGACUUUCACUCAA_AGUGAAAGUUUGCAUUU_CUUAGCUUUCGCUC_UCGAUUUCCAA___UUGCCAAAAAUUGUUGCUUAGCCG ..........(((((((((((....)))))))))))(((............(((.....)))...........))).................... (-10.94 = -11.05 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:57 2006