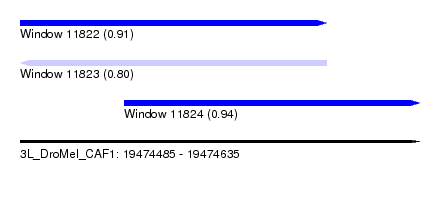
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,474,485 – 19,474,635 |
| Length | 150 |
| Max. P | 0.944868 |

| Location | 19,474,485 – 19,474,600 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.76 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19474485 115 + 23771897 AAGUUCAAUUGGUGUGACCUGCCAAUUGGCAUAAAGGAAAAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCU-GUGAAA-AGUUUCCGGCGAAAAUGUCGGGAAAAUACG ..((.((((((((.......)))))))))).............(((((...(((((((.....)))........)))).-.)))))-..((((((((......))))))))...... ( -33.30) >DroSec_CAF1 114461 116 + 1 AAGUUCAAUUGGUGUGACCUGCCAAUUGGCAUAAAGGAAAAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCUCGUGGAA-AGUUUCCGGGGAAAAUGUCGGGGAAAUACG ...........((((..((((((....))).....(((((...(((((...(((((((.....)))........))))...)))))-..)))))............)))...)))). ( -31.30) >DroSim_CAF1 114582 116 + 1 AAGUUCAAUUGGUGUGACCUGCCAAUUGGCAUAAAGGAAAAGAUUUCAAAUAGCCGCCAAGUUGGCAAAAUGUUGGCCUCGCGGAA-AGUUUCCGGGGAAAAUGUCGGGGAAAUACG ...........((((..(((((((((((((.((...(((.....)))...))...))).)))))))...(((((..(((((.(((.-...))))))))..))))).)))...)))). ( -30.50) >DroEre_CAF1 99944 113 + 1 AAGUUCAAUUGGUGUGACCUGCCAAUUGGCAUAAAGGAAAAGAUUUCGAAUGGCCGCCAAGUUGCCAAAAUGUUGGCCU-GUGGAA-AUUUUCCGGGGAAAAUGCCGGGC--GUACG ..(((((((((((.......)))))))(((((...((((((..(((((...(((((....(((.....)))..))))).-.)))))-.)))))).......)))))))))--..... ( -33.40) >DroYak_CAF1 152973 114 + 1 AAGUUCAAUUGGUGUGACCUGCCAAUUGGCAUAAAGGAAAAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCU-GUGGAAAAGUUUCCGGGAAAAAUGCCGGGC--GUACG ..(((((((((((.......)))))))(((((...(((((...(((((...(((((((.....)))........)))).-.)))))...))))).......)))))))))--..... ( -35.40) >consensus AAGUUCAAUUGGUGUGACCUGCCAAUUGGCAUAAAGGAAAAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCU_GUGGAA_AGUUUCCGGGGAAAAUGUCGGGCAAAUACG ..((....((((..(..((((((....))).....(((((...(((((...(((((((.....)))........))))...)))))...))))))))....)..))))......)). (-28.48 = -28.76 + 0.28)


| Location | 19,474,485 – 19,474,600 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.04 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19474485 115 - 23771897 CGUAUUUUCCCGACAUUUUCGCCGGAAACU-UUUCAC-AGGCCAACAUUUUGCCAACUUGGCGGCCAUUUGAAAUCUUUUCCUUUAUGCCAAUUGGCAGGUCACACCAAUUGAACUU ...........(((.........(((((..-(((((.-.((((........(((.....)))))))...)))))...)))))....((((....)))).)))............... ( -26.80) >DroSec_CAF1 114461 116 - 1 CGUAUUUCCCCGACAUUUUCCCCGGAAACU-UUCCACGAGGCCAACAUUUUGCCAACUUGGCGGCCAUUUGAAAUCUUUUCCUUUAUGCCAAUUGGCAGGUCACACCAAUUGAACUU ...........(((.........(((((.(-(((.....((((........(((.....)))))))....))))...)))))....((((....)))).)))............... ( -23.70) >DroSim_CAF1 114582 116 - 1 CGUAUUUCCCCGACAUUUUCCCCGGAAACU-UUCCGCGAGGCCAACAUUUUGCCAACUUGGCGGCUAUUUGAAAUCUUUUCCUUUAUGCCAAUUGGCAGGUCACACCAAUUGAACUU ...........(((.....(((((((....-.)))).).))........(((((((.((((((.......(((.....))).....))))))))))))))))............... ( -25.30) >DroEre_CAF1 99944 113 - 1 CGUAC--GCCCGGCAUUUUCCCCGGAAAAU-UUCCAC-AGGCCAACAUUUUGGCAACUUGGCGGCCAUUCGAAAUCUUUUCCUUUAUGCCAAUUGGCAGGUCACACCAAUUGAACUU .(..(--(((.((.((((((....))))))-..))..-..(((((....))))).....))))..).(((((..............((((....))))((.....))..)))))... ( -29.30) >DroYak_CAF1 152973 114 - 1 CGUAC--GCCCGGCAUUUUUCCCGGAAACUUUUCCAC-AGGCCAACAUUUUGCCAACUUGGCGGCCAUUUGAAAUCUUUUCCUUUAUGCCAAUUGGCAGGUCACACCAAUUGAACUU .....--(((.(((((.......(((((..((((...-.((((........(((.....)))))))....))))...)))))...)))))....))).((.....)).......... ( -26.90) >consensus CGUAUUUCCCCGACAUUUUCCCCGGAAACU_UUCCAC_AGGCCAACAUUUUGCCAACUUGGCGGCCAUUUGAAAUCUUUUCCUUUAUGCCAAUUGGCAGGUCACACCAAUUGAACUU ...........(((.........(((((...(((.....((((........(((.....)))))))....)))....)))))....((((....)))).)))............... (-21.40 = -21.04 + -0.36)


| Location | 19,474,524 – 19,474,635 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19474524 111 + 23771897 AAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCU-GUGAAA-AGUUUCCGGCGAAAAUGUCGGGAAAAUACGGUCGUGAAAUGAAAAGUGGAGGCUAUGAAAAGGGC ....(((((..(((((.(((.((..((.....))..))((-(((...-..((((((((......))))))))..)))))...............))).))))))))))..... ( -32.10) >DroSec_CAF1 114500 112 + 1 AAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCUCGUGGAA-AGUUUCCGGGGAAAAUGUCGGGGAAAUACGGUCGUGAAAUGAAAAGUGGAGGCUAUGAAAAGGGC ....(((((..(((((.(((.((..((.....))..)).(((((...-..(..((((........))))..)..)))))...............))).))))))))))..... ( -29.20) >DroSim_CAF1 114621 112 + 1 AAGAUUUCAAAUAGCCGCCAAGUUGGCAAAAUGUUGGCCUCGCGGAA-AGUUUCCGGGGAAAAUGUCGGGGAAAUACGGUCGCGAAAUGAAAAGUGGAGGCUAUGAAAAGGGC ....(((((..(((((.(((.((..((.....))..)).((((((..-..(..((((........))))..).......)))))).........))).))))))))))..... ( -34.30) >DroEre_CAF1 99983 109 + 1 AAGAUUUCGAAUGGCCGCCAAGUUGCCAAAAUGUUGGCCU-GUGGAA-AUUUUCCGGGGAAAAUGCCGGGC--GUACGGUUGUGAAAUGAAAAGUGGAGGCUAUGAAAAGGGC ....((((..((((((.(((..(((((((....)))))((-.((((.-....)))).)).....((((...--...))))..........))..))).))))))))))..... ( -29.90) >DroYak_CAF1 153012 110 + 1 AAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCU-GUGGAAAAGUUUCCGGGAAAAAUGCCGGGC--GUACGGUCGUGAAAUGAAAAGUGGAGGCUAUGAAAAGGGC ....(((((.((((((((((.((..((.....))..))..-.))).......(((((........))))).--...)))))))....))))).......(((.(....).))) ( -28.20) >consensus AAGAUUUCAAAUGGCCGCCAAGUUGGCAAAAUGUUGGCCU_GUGGAA_AGUUUCCGGGGAAAAUGUCGGGCAAAUACGGUCGUGAAAUGAAAAGUGGAGGCUAUGAAAAGGGC ....(((((..(((((.(((..((..((......(((((..(((......(((((((........)))))))..)))))))).....)).))..))).))))))))))..... (-24.84 = -25.04 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:49 2006