
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,455,284 – 19,455,375 |
| Length | 91 |
| Max. P | 0.873090 |

| Location | 19,455,284 – 19,455,375 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19455284 91 + 23771897 AGCGCCGGCAUCCUGCCACUUUGUUUGCCGGC-AAACAUUUGUGCAGCUUAUUAAUAUGCAUGCCACCCGGCUCUUCUC------------------CCCCGGCUCCUCA ...(((((....((((.((..((((((....)-)))))...))))))...............(((....))).......------------------..)))))...... ( -26.00) >DroSec_CAF1 96812 90 + 1 AGCGCCGGCAUCCUGCCACUUUGUUUGCCGGC-AAACAUUUGUGCAGCUUAUUAAUAUGCAUGCCACCCGGCUCUUCUC------------------C-CCGGCACCUCA ...(((((....((((.((..((((((....)-)))))...))))))...............(((....))).......------------------.-)))))...... ( -26.50) >DroSim_CAF1 97474 92 + 1 AGCGCCGGCAUCCUGCCACUUUGUUUGCCAUCUAGAGAUUUGUGCAGCUUAUUAAUAUGCAUGCCACCCGGCUCUUCUC------------------CCCCGGCUCCUCA ...(((((((..(.........)..)))).....((((...(((((...........)))))(((....)))...))))------------------....)))...... ( -23.50) >DroEre_CAF1 84004 109 + 1 AGCGCCGGCAUCCUGCCACUUUGUUUGCCGGC-AAACAUUUGUGCAGCUUAUUAAUAUGCAUGCCACCCGACUCUGCUCCCCCGGCUCCCCCGACUGUCCCGGCUCCUCA ((((((((((..(.........)..)))))))-.......((.(((((..........)).))))).........)))...((((..(........)..))))....... ( -27.10) >DroYak_CAF1 136493 99 + 1 AGCGCCGGCAUCCUGCCACUUUGUUUGCUGGC-AAACAUUUGUGCAGCUUAUUAAUAUGCAUGCCACCCGAC------CACCCGGC----CCUGCUCCCCCGGCUCCUCA ...(((((....((((.((..((((((....)-)))))...))))))...........(((.(((.......------.....)))----..)))....)))))...... ( -27.40) >DroAna_CAF1 84146 78 + 1 AGCCCCGGCAUCCUGGCACUUUGUUUGCCGGC-AAACAUUUGUGCAGCUUAUUAAUAUGCACGCCCCGCACCU-------------------------------UCCUCG .((...(((......((((..((((((....)-)))))...)))).((..........))..)))..))....-------------------------------...... ( -20.70) >consensus AGCGCCGGCAUCCUGCCACUUUGUUUGCCGGC_AAACAUUUGUGCAGCUUAUUAAUAUGCAUGCCACCCGGCUCUUCUC__________________CCCCGGCUCCUCA .(((((((((..(.........)..))))))).........(((((...........))))))).............................................. (-17.04 = -17.27 + 0.22)

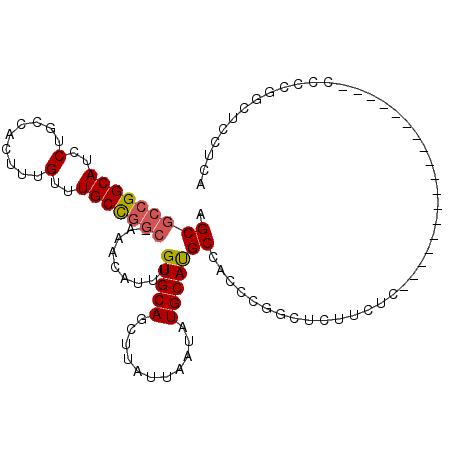
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:38 2006