| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
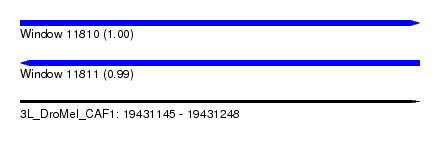
| Location | 19,431,145 – 19,431,248 |
| Length | 103 |
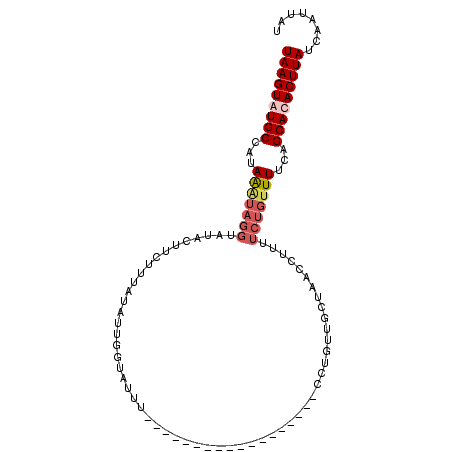
| Max. P | 0.997908 |

| Location | 19,431,145 – 19,431,248 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 66.72 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -8.31 |
| Energy contribution | -9.74 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19431145 103 + 23771897 UAAGUGUGCCAUAAGUAGGUAGACAUUUUAUUAAUGGUAUUUACAUACAUAUAUACAUUUCUCGUUCCUAACCUUUUCUGUUUUCAGCACACUUAUCAAUUAU (((((((((....((.((((((.((((.....))))((((....))))...................)).))))...)).......)))))))))........ ( -18.70) >DroSec_CAF1 70045 85 + 1 UAAGUAUGCCAUAAAUAGGUAUACUUCUUUAUAUUGGUAUUU------------------CCUUUUGCUAACCUUUUCUGUUUUUAGCACACUUAUUAAAACU (((((.(((...(((((((((((......))))((((((...------------------.....)))))).....)))))))...))).)))))........ ( -14.00) >DroSim_CAF1 72378 85 + 1 UAAGUAUGCCAUAAAUAGGUAUACUUCUUUAUAUUGGUAUUU------------------CCUGUUGCUAACCUUUUCUGUUUUCAGCACACUUAUCAAUUAC .(((((((((.......)))))))))......(((((((...------------------..(((((..(((.......)))..)))))....)))))))... ( -18.50) >DroYak_CAF1 112516 87 + 1 UAAGUGUGCCACAGAUAGGUAUCAUGCU---UAUUGGUAGCU----ACUUAUAUGCACUUUCAGUCUCUAACUUUA-----UUUCAGCACACUUACUAA---- (((((((((..((.(((((((...((((---....))))..)----)))))).)).......(((.....)))...-----.....)))))))))....---- ( -19.60) >consensus UAAGUAUGCCAUAAAUAGGUAUACUUCUUUAUAUUGGUAUUU__________________CCUGUUGCUAACCUUUUCUGUUUUCAGCACACUUAUCAAUUAU (((((((((...(((((((.........................................................)))))))...)))))))))........ ( -8.31 = -9.74 + 1.44)


| Location | 19,431,145 – 19,431,248 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 66.72 |
| Mean single sequence MFE | -16.86 |
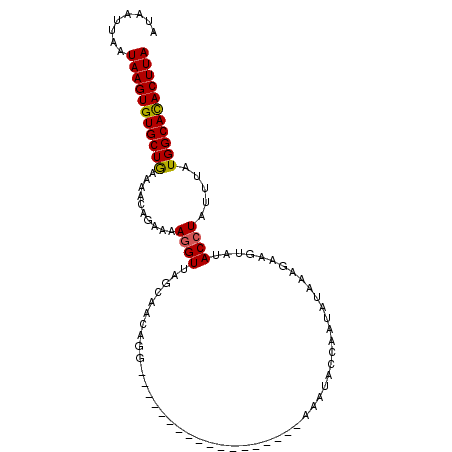
| Consensus MFE | -9.68 |
| Energy contribution | -9.49 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19431145 103 - 23771897 AUAAUUGAUAAGUGUGCUGAAAACAGAAAAGGUUAGGAACGAGAAAUGUAUAUAUGUAUGUAAAUACCAUUAAUAAAAUGUCUACCUACUUAUGGCACACUUA ........(((((((((((....).....((((.(((....(((...((((.(((....))).))))((((.....))))))).)))))))..)))))))))) ( -18.40) >DroSec_CAF1 70045 85 - 1 AGUUUUAAUAAGUGUGCUAAAAACAGAAAAGGUUAGCAAAAGG------------------AAAUACCAAUAUAAAGAAGUAUACCUAUUUAUGGCAUACUUA ........(((((((((((.(((.((....(((...(.....)------------------....))).((((......))))..)).))).))))))))))) ( -15.60) >DroSim_CAF1 72378 85 - 1 GUAAUUGAUAAGUGUGCUGAAAACAGAAAAGGUUAGCAACAGG------------------AAAUACCAAUAUAAAGAAGUAUACCUAUUUAUGGCAUACUUA ........(((((((((((.(((.((....(((...(.....)------------------....))).((((......))))..)).))).))))))))))) ( -14.30) >DroYak_CAF1 112516 87 - 1 ----UUAGUAAGUGUGCUGAAA-----UAAAGUUAGAGACUGAAAGUGCAUAUAAGU----AGCUACCAAUA---AGCAUGAUACCUAUCUGUGGCACACUUA ----....((((((((((....-----....(.(((..(((.............)))----..))).)....---.(((.(((....)))))))))))))))) ( -19.12) >consensus AUAAUUAAUAAGUGUGCUGAAAACAGAAAAGGUUAGCAACAGG__________________AAAUACCAAUAUAAAGAAGUAUACCUAUUUAUGGCACACUUA ........(((((((((((..........((((..................................................)))).....))))))))))) ( -9.68 = -9.49 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:34 2006