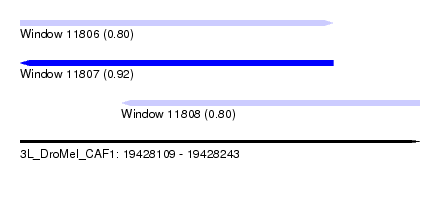
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,428,109 – 19,428,243 |
| Length | 134 |
| Max. P | 0.916140 |

| Location | 19,428,109 – 19,428,214 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -17.97 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19428109 105 + 23771897 GGCAAAGAGACAGAUGGCGGGGCGAACUU-----CAAGCGCAAUUUUACGCAACAACAAAAAGUUUAAUUGAGGAUAAACUUUGCUGCGUUUGCGCUGCUGUUGAAAAUC--------- ............((((((((.((((((..-----..(((((........))........((((((((........)))))))))))..)))))).)))))))).......--------- ( -28.50) >DroVir_CAF1 24694 105 + 1 GGCAC--AGACAAA---CUGAACGAACCCCAACUCUGGCGCAAUUUUACGCAACAACAAAAAGUUUAAUUGAGGAUAAACUUUGCUGCGUUUGCGCUGCCGUUGAAAAUC--------- (((.(--((.....---)))................(((((((....(((((.(((.....((((((........))))))))).)))))))))))))))..........--------- ( -26.00) >DroPse_CAF1 34200 102 + 1 --CACAUAGACGGAUG-UGGACCCAACUU-----CAAGCGCAAUUUUCCGCAACAACAAAAAGUUUAAUUGAGGAUAAACUUUGCUGCGUUUGCGCUGCUGUUGAAAAUC--------- --(((((......)))-))....((((..-----..(((((((.....((((.(((.....((((((........))))))))).)))).)))))))...))))......--------- ( -26.20) >DroWil_CAF1 101048 105 + 1 --CACUCCCCCAAAAA----A---CACAU-----UUUGCGCAAUUUUACGCAACAACAAAAAGUUUAAUUGAGGAUAAACUUUGCUGCGUUUGCGCUGCUGUUGAAAAUUCUACCCCAC --..............----.---.....-----.(((((........)))))(((((.((((((((........))))))))((.(((....))).)))))))............... ( -20.20) >DroAna_CAF1 61340 105 + 1 UUCACUCACACAAAGACUGAGGCCAACUU-----CAAGCGCAAUUUUACGCAACAACAAAAAGUUUAAUUGAGGAUAAACUUUGCUGCGUUUGCGCUGCUGUUGAAAAUC--------- ....((((.........))))..((((..-----..(((((((....(((((.(((.....((((((........))))))))).))))))))))))...))))......--------- ( -26.00) >DroPer_CAF1 34190 102 + 1 --CACAUAGACGGAUG-UGGACCCAACUU-----CAAGCGCAAUUUUCCGCAACAACAAAAAGUUUAAUUGAGGAUAAACUUUGCUGCGUUUGCGCUGCUGUUGAAAAUC--------- --(((((......)))-))....((((..-----..(((((((.....((((.(((.....((((((........))))))))).)))).)))))))...))))......--------- ( -26.20) >consensus __CACACAGACAAAUG_UGGACCCAACUU_____CAAGCGCAAUUUUACGCAACAACAAAAAGUUUAAUUGAGGAUAAACUUUGCUGCGUUUGCGCUGCUGUUGAAAAUC_________ .....................................(((........)))..(((((.((((((((........))))))))((.(((....))).)))))))............... (-17.97 = -18.13 + 0.17)



| Location | 19,428,109 – 19,428,214 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19428109 105 - 23771897 ---------GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGCGCUUG-----AAGUUCGCCCCGCCAUCUGUCUCUUUGCC ---------((.(((((.....((((((((((((((((((((((........)))))).....)))))))))....)))))))))-----))).))....................... ( -27.20) >DroVir_CAF1 24694 105 - 1 ---------GAUUUUCAACGGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGCGCCAGAGUUGGGGUUCGUUCAG---UUUGUCU--GUGCC ---------((.(..((((....(((((((((((((((((((((........)))))).....)))))))))....))))))....))))..).))((.(((---.....))--).)). ( -34.20) >DroPse_CAF1 34200 102 - 1 ---------GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGGAAAAUUGCGCUUG-----AAGUUGGGUCCA-CAUCCGUCUAUGUG-- ---------....((((((..((((((((.((((((((((((((........)))))).....)))))))).....)))))).))-----..))))))..((-(((......)))))-- ( -30.40) >DroWil_CAF1 101048 105 - 1 GUGGGGUAGAAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGCGCAAA-----AUGUG---U----UUUUUGGGGGAGUG-- .((..(......)..))(((...(((((((((((((((((((((........)))))).....)))))))))....))))))...-----.))).---.----..............-- ( -26.00) >DroAna_CAF1 61340 105 - 1 ---------GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGCGCUUG-----AAGUUGGCCUCAGUCUUUGUGUGAGUGAA ---------.....(((((..(((((((((((((((((((((((........)))))).....)))))))))....)))))).))-----..)))))..(((.((.......)).))). ( -32.40) >DroPer_CAF1 34190 102 - 1 ---------GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGGAAAAUUGCGCUUG-----AAGUUGGGUCCA-CAUCCGUCUAUGUG-- ---------....((((((..((((((((.((((((((((((((........)))))).....)))))))).....)))))).))-----..))))))..((-(((......)))))-- ( -30.40) >consensus _________GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGCGCUUG_____AAGUUGGCUCCA_CAUCUGUCUAUGUG__ .......................(((((((((((((((((((((........)))))).....)))))))))....))))))..................................... (-22.05 = -22.38 + 0.33)

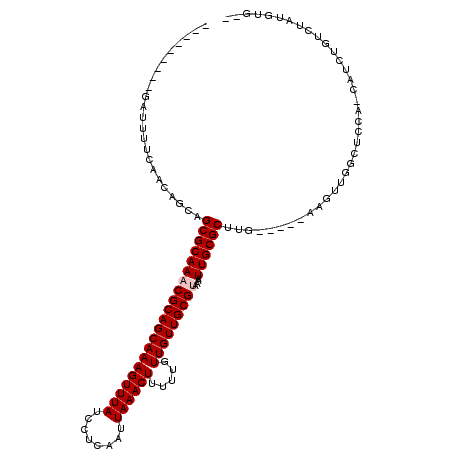
| Location | 19,428,143 – 19,428,243 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.88 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19428143 100 - 23771897 --------UGG--GGAAAUUUUCCAUGUCGACGUUGGUU----------GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGC --------...--(((((((..(((((....)).)))..----------)))))))....(((((((((..((((.((((((((........))))))))))))..)))))).....))) ( -26.00) >DroVir_CAF1 24728 93 - 1 -----------------CUUUUCUAUCCGGCCCGCAGUU----------GAUUUUCAACGGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGC -----------------............((..((.(((----------(.....)))).)).))(((((((((((((((((((........)))))).....)))))))))....)))) ( -24.30) >DroSec_CAF1 66675 100 - 1 --------UUG--GGAAAUUUUUCCUGGCGACGUUGGUU----------GAUUUUCAACAGCCACGCAAACGCAGCAAAGUUUAUCCUCCAUUUAACUUUUUGUUGUUGCGUGAAAUUGC --------..(--((((....))))).((((.((((.((----------(.....)))))))(((((((..((((.((((((............))))))))))..)))))))...)))) ( -25.60) >DroWil_CAF1 101073 97 - 1 -----------------------CAUUCCGGCGUAGUUUUGUGGGGUAGAAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGC -----------------------.......((((.(((...((..(......)..))..))).))))..(((((((((((((((........)))))).....)))))))))........ ( -25.30) >DroAna_CAF1 61374 110 - 1 UGGGUAGUUGGGGGAGAAUUUUCCAUGUCGACGUUGGUU----------GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGC ((.((.((((...(((((((..(((((....)).)))..----------)))))))..)))).)).)).(((((((((((((((........)))))).....)))))))))........ ( -33.20) >DroPer_CAF1 34221 98 - 1 --------UGG----GAAUUUUCCAUGUCGACGUUGGUU----------GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGGAAAAUUGC --------..(----.((((((((..((((((....)))----------)))...(((((((((((....)))...((((((((........)))))))).)))))))).)))))))).) ( -30.00) >consensus ________UGG_____AAUUUUCCAUGUCGACGUUGGUU__________GAUUUUCAACAGCAGCGCAAACGCAGCAAAGUUUAUCCUCAAUUAAACUUUUUGUUGUUGCGUAAAAUUGC .................................................(((((((((((((((((....)))...((((((((........)))))))).))))))))...)))))).. (-15.80 = -15.88 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:32 2006