
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,419,633 – 19,419,727 |
| Length | 94 |
| Max. P | 0.987386 |

| Location | 19,419,633 – 19,419,727 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
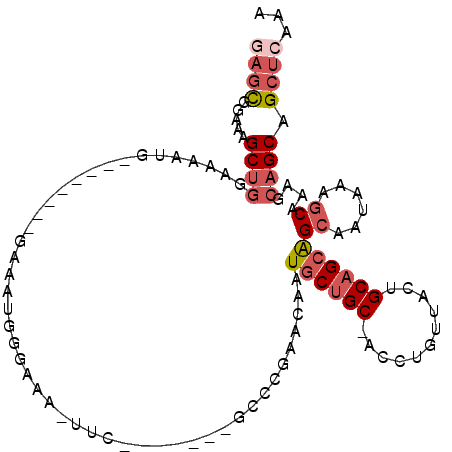
>3L_DroMel_CAF1 19419633 94 + 23771897 UUUGAGCUGCUGCUUUGCUUUAUUGCUGCUGCAGUAACAGGU-GCAGCAUUGUUCGUUC-------GAA-UUUCCCAUUUUCCAG------CUUUCCAGCUUUCCGCUC ...((((((((((((((..(((((((....))))))))))).-))))))..((((....-------)))-)............((------((....))))....)))) ( -24.90) >DroVir_CAF1 16525 91 + 1 UUGGAGCUGCUGUUUUGGUUUAUUGCCGCUGCAGCAGACGGUCGCUG-AUGGUGCUGCCGGCCAGGGGAUAAU----------------CAGGGACGGGCCUGAUAAU- ((((..(((((((..((((.....))))..))))))).((((.((..-.....)).)))).))))......((----------------((((......))))))...- ( -35.30) >DroSec_CAF1 58378 93 + 1 UUUGAGCUGCUGCUUUGCUUUAUUGCUGCUGCAGUAACAGGU-GCAGCAUUGUUCGGGC-------CAA-UUUCCCAUUUC-------CCAUUUUCCAGCUUUCCGCUA ...((((((((((((((..(((((((....))))))))))).-))))))..))))(((.-------...-...))).....-------.........(((.....))). ( -26.30) >DroSim_CAF1 59550 100 + 1 UUUGAGCUGCUGCUUUGCUUUAUUGCUGCUGCAGUAACAGGU-GCAGCAUUGUUCGGGC-------GAA-UUUCCCAUUUCCCAGUUCCCAUUUUCCAGCUUUCCGCUC ...((((((((((((((..(((((((....))))))))))).-))))))..))))(((.-------(((-(.....)))))))..............(((.....))). ( -26.60) >DroEre_CAF1 54830 93 + 1 UUUGAGCUGCUGCUUUGCUUUAUUGCUGCUGCAGUAACAGGU-GCAGCAUUGUUCGGGG-------GAA-UUUCCCAUUUC-------CCAUUUUCCAGCUUUCUGCGC .((((((((((((((((..(((((((....))))))))))).-))))))..))))))((-------(((-........)))-------))........((.....)).. ( -30.80) >DroYak_CAF1 98453 86 + 1 UUUGAGCUGCUGCUUUGCUUUAUUGCUGCUGCAGUAACAGGU-GCAGCAUUGUUCGGGG-------GAA-UUUCC--------------CAGUUUCCAGCUUUCCACUC ...((((((((((((((..(((((((....))))))))))).-))))))..))))((((-------(..-...))--------------)(((.....)))..)).... ( -27.70) >consensus UUUGAGCUGCUGCUUUGCUUUAUUGCUGCUGCAGUAACAGGU_GCAGCAUUGUUCGGGC_______GAA_UUUCCCAUUUC________CAUUUUCCAGCUUUCCGCUC ..(((((((((((...((((((((((....)))))))..))).))))))..)))))..................................................... (-17.22 = -17.50 + 0.28)



| Location | 19,419,633 – 19,419,727 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.62 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19419633 94 - 23771897 GAGCGGAAAGCUGGAAAG------CUGGAAAAUGGGAAA-UUC-------GAACGAACAAUGCUGC-ACCUGUUACUGCAGCAGCAAUAAAGCAAAGCAGCAGCUCAAA ((((....((((....))------)).............-...-------..........((((((-..((((....))))..((......))...))))))))))... ( -27.30) >DroVir_CAF1 16525 91 - 1 -AUUAUCAGGCCCGUCCCUG----------------AUUAUCCCCUGGCCGGCAGCACCAU-CAGCGACCGUCUGCUGCAGCGGCAAUAAACCAAAACAGCAGCUCCAA -...((((((......))))----------------)).......(((.(((..((.....-..))..))).((((((....((.......))....))))))..))). ( -22.30) >DroSec_CAF1 58378 93 - 1 UAGCGGAAAGCUGGAAAAUGG-------GAAAUGGGAAA-UUG-------GCCCGAACAAUGCUGC-ACCUGUUACUGCAGCAGCAAUAAAGCAAAGCAGCAGCUCAAA ((((.....)))).....(((-------(...((((...-...-------.)))).....((((((-..((((....))))..((......))...)))))).)))).. ( -25.40) >DroSim_CAF1 59550 100 - 1 GAGCGGAAAGCUGGAAAAUGGGAACUGGGAAAUGGGAAA-UUC-------GCCCGAACAAUGCUGC-ACCUGUUACUGCAGCAGCAAUAAAGCAAAGCAGCAGCUCAAA ((((.....((((...................((((...-...-------.)))).....((((((-(........)))))))((......))....)))).))))... ( -26.60) >DroEre_CAF1 54830 93 - 1 GCGCAGAAAGCUGGAAAAUGG-------GAAAUGGGAAA-UUC-------CCCCGAACAAUGCUGC-ACCUGUUACUGCAGCAGCAAUAAAGCAAAGCAGCAGCUCAAA ...(((....))).....(((-------(...((((...-...-------.)))).....((((((-..((((....))))..((......))...)))))).)))).. ( -23.60) >DroYak_CAF1 98453 86 - 1 GAGUGGAAAGCUGGAAACUG--------------GGAAA-UUC-------CCCCGAACAAUGCUGC-ACCUGUUACUGCAGCAGCAAUAAAGCAAAGCAGCAGCUCAAA ((((((......(....).(--------------((...-..)-------))))......((((((-..((((....))))..((......))...))))))))))... ( -26.30) >consensus GAGCGGAAAGCUGGAAAAUG________GAAAUGGGAAA_UUC_______GCCCGAACAAUGCUGC_ACCUGUUACUGCAGCAGCAAUAAAGCAAAGCAGCAGCUCAAA ((((.....((((...............................................((((((...........))))))((......))....)))).))))... (-11.48 = -12.62 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:25 2006