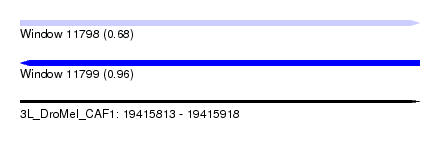
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,415,813 – 19,415,918 |
| Length | 105 |
| Max. P | 0.964585 |

| Location | 19,415,813 – 19,415,918 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19415813 105 + 23771897 AAAUUGCACAUGAGCUGCUGU---UGCAUCUUGAUUGCAC-----UUAUCCUGGGUGUACCCAUAAAUGCAUGCAUGAGCAAGGACAUCCUCGUAUGCACAUAAGUAUGCGUU ....((((..((((.(((...---.))).))))..)))).-----......((((....))))..((((((((((((.((((((....)))....))).)))..))))))))) ( -29.70) >DroSec_CAF1 54548 108 + 1 AAAUGGCACACGAGCUGCAGUAGCUGCAUCUUGAUUGCAC-----UUUUCCUGGGUGUACCCAUAAAUGCAUGCAUGAGCAAGGACAUCCUCGUAUGCACAUAAGUAUGCGUU ..((((....((((.(((((...))))).))))..(((((-----((.....))))))).)))).((((((((((((.((((((....)))....))).)))..))))))))) ( -34.10) >DroSim_CAF1 55666 108 + 1 AAAUUGCACAUGAGCUGCUGUAGCUGCAUCUUGAUUGCAC-----UUUUCCUGGGUGUACCCAUAAAUGCAUGCAUGAGCAAGGACAUCCUCGUAUGCACAUAAGUAUGCGUU ...((((.(((((((((...)))))((((..((..(((((-----((.....)))))))..))...))))...)))).))))((....)).(((((((......))))))).. ( -31.60) >DroEre_CAF1 51032 110 + 1 AAAUUAAAUGUGGAUUGCUGU---UGCAUCUCGAUUGCAUCUAGAUUUUCCUGGGUGUACUCAUAAAUGCAUGCAUGAGCAAGGACAUCCUCAUAUGCACGUAAGUAUGCGUU ...........((..(((...---.)))..))((.(((((((((......))))))))).))...((((((((((((.((((((....)))....))).)))..))))))))) ( -31.60) >DroYak_CAF1 80979 97 + 1 GACUUAAAUGUGAAUUGCUGU---AGCAUCUAGAUUGCAC-----UUUUCCUGUGUGUGCUCAUAAAUGCAUGCAUGAGCAAGGACA--------UGCACGUAAGUAUGUGUU .......((((...(((((..---.(((.......)))..-----.......((((((((........)))))))).)))))..)))--------)((((((....)))))). ( -24.10) >consensus AAAUUGCACAUGAGCUGCUGU___UGCAUCUUGAUUGCAC_____UUUUCCUGGGUGUACCCAUAAAUGCAUGCAUGAGCAAGGACAUCCUCGUAUGCACAUAAGUAUGCGUU ..........((((.(((.......))).))))..................((((....))))..((((((((((((.((((((....)))....))).)))..))))))))) (-20.32 = -20.76 + 0.44)

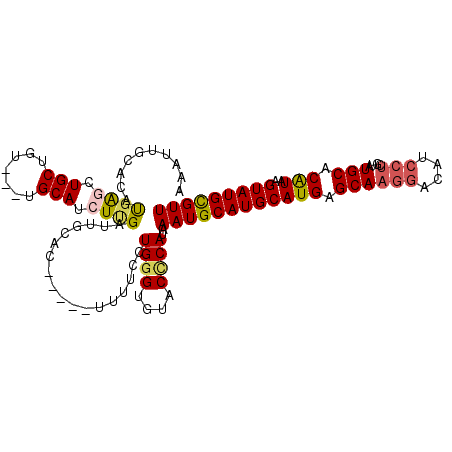

| Location | 19,415,813 – 19,415,918 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.21 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19415813 105 - 23771897 AACGCAUACUUAUGUGCAUACGAGGAUGUCCUUGCUCAUGCAUGCAUUUAUGGGUACACCCAGGAUAA-----GUGCAAUCAAGAUGCA---ACAGCAGCUCAUGUGCAAUUU ...((((....))))(((((.(((..(((..((((((.((..((((((((((((....))))...)))-----)))))..)).)).)))---)..))).))).)))))..... ( -33.30) >DroSec_CAF1 54548 108 - 1 AACGCAUACUUAUGUGCAUACGAGGAUGUCCUUGCUCAUGCAUGCAUUUAUGGGUACACCCAGGAAAA-----GUGCAAUCAAGAUGCAGCUACUGCAGCUCGUGUGCCAUUU ...........(((.((((((((((((.....(((....)))(((((((.((((....))))....))-----))))))))....(((((...))))).)))))))))))).. ( -36.80) >DroSim_CAF1 55666 108 - 1 AACGCAUACUUAUGUGCAUACGAGGAUGUCCUUGCUCAUGCAUGCAUUUAUGGGUACACCCAGGAAAA-----GUGCAAUCAAGAUGCAGCUACAGCAGCUCAUGUGCAAUUU ...((((.(((..((((((.(((((....)))))...))))))((((((.((((....))))....))-----))))....)))(((.((((.....)))))))))))..... ( -33.10) >DroEre_CAF1 51032 110 - 1 AACGCAUACUUACGUGCAUAUGAGGAUGUCCUUGCUCAUGCAUGCAUUUAUGAGUACACCCAGGAAAAUCUAGAUGCAAUCGAGAUGCA---ACAGCAAUCCACAUUUAAUUU ...((((.(((...(((((...(((...((((((((((((........)))))))).....))))...)))..)))))...))))))).---..................... ( -26.40) >DroYak_CAF1 80979 97 - 1 AACACAUACUUACGUGCA--------UGUCCUUGCUCAUGCAUGCAUUUAUGAGCACACACAGGAAAA-----GUGCAAUCUAGAUGCU---ACAGCAAUUCACAUUUAAGUC .......(((((..((((--------(.((((((((((((........)))))))).....))))...-----))))).......(((.---...))).........))))). ( -23.30) >consensus AACGCAUACUUAUGUGCAUACGAGGAUGUCCUUGCUCAUGCAUGCAUUUAUGGGUACACCCAGGAAAA_____GUGCAAUCAAGAUGCA___ACAGCAGCUCAUGUGCAAUUU ...((((.(((...(((((.........((((((((((((........)))))))).....))))........)))))...)))))))......................... (-21.25 = -21.21 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:23 2006