
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,414,552 – 19,414,792 |
| Length | 240 |
| Max. P | 0.999020 |

| Location | 19,414,552 – 19,414,672 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -32.26 |
| Energy contribution | -33.38 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19414552 120 + 23771897 GGACUGGAAUCCGCACAACGACGUUCAGGCCUUUUCCCGAGCCCAUCGCAUGGGGCAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCA (((......)))(((.((.(.(((((..(((((((.....((((........))))......))))))).....((((((((.(((((...))))).)))))))).)))))).)).))). ( -41.90) >DroSec_CAF1 53324 120 + 1 GGACUGGAAUCCGCACAACGACGUUCAGGCCUUCUCCCGAGCCCAUCGCAUGGGACAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCA (((......)))(((.((.(.(((((..(((((((((((.((.....)).)))))........)))))).....((((((((.(((((...))))).)))))))).)))))).)).))). ( -43.20) >DroSim_CAF1 54409 120 + 1 GGACUGGAAUCCGCACAACGACGUUCAGGCCUUCUCCCGAGCCCAUCGCAUGGGACAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACUCACAACUCGGUGGAGGAGCGCAUUAUGCA (((......)))(((.((.(.(((((..(((((((((((.((.....)).)))))........)))))).....((((((((.(((((...))))).)))))))).)))))).)).))). ( -43.20) >DroEre_CAF1 49770 120 + 1 CGACUGGAAUCCACACAACGACGUUCAGGCCUUCUCACGAGCCCAUCGCAUGGGUCAGACAAAAAAGGUGAUGAUCUACCGAUUUGUGACCCACAACUCGGUCGAGGAGCGCAUGAUGCA ...(((((.((........))..)))))((((((((.(((((.....)).((((((..(((((...((((......))))..)))))))))))....)))...)))))).))........ ( -33.10) >DroYak_CAF1 79737 120 + 1 CGACUGGAAUCCACACAACGACGUUCAGGCCUUCUCAAGAGCUCAUCGCAUGGGUCAGACAAAGAAGGUGAUGAUCUACCGAUUUGUGACCCACAACUCCGUGGAGGAGCGCAUCAUGCA .(((((((.((........))..)))))((((((((....((.....)).((((((..(((((...((((......))))..)))))))))))..........)))))).)).))..... ( -32.90) >consensus GGACUGGAAUCCGCACAACGACGUUCAGGCCUUCUCCCGAGCCCAUCGCAUGGGACAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCA ............(((.((.(.(((((..(((((((......(((((...)))))........))))))).....((((((((.(((((...))))).)))))))).)))))).)).))). (-32.26 = -33.38 + 1.12)


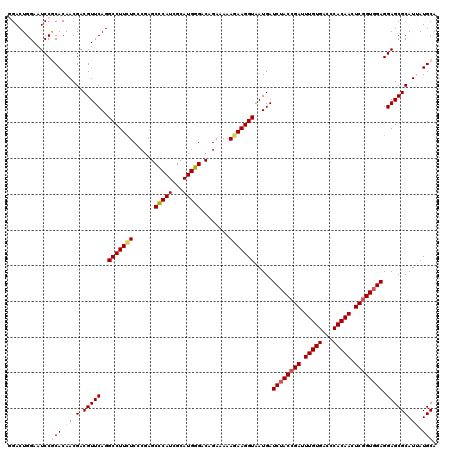
| Location | 19,414,592 – 19,414,712 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -38.10 |
| Energy contribution | -37.70 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19414592 120 + 23771897 GCCCAUCGCAUGGGGCAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCACAAGAUGAUGCUCACCCAUCUUGUGGUG ((((........))))..........((((((..((((((((.(((((...))))).)))))))).....((.....))..))))))...(((((((((.........)))))))))... ( -44.00) >DroSec_CAF1 53364 120 + 1 GCCCAUCGCAUGGGACAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCGCAAGAUGAUGCUCACCCAUCUUGUGGUG ...(((((((((((....................((((((((.(((((...))))).)))))))).(((((((((.(((..(((....)))))).)))).))))).))))...))))))) ( -43.20) >DroSim_CAF1 54449 120 + 1 GCCCAUCGCAUGGGACAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACUCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCGCAAGAUGAUGCUCACCCAUCUUGUGGUG ...(((((((((((....................((((((((.(((((...))))).)))))))).(((((((((.(((..(((....)))))).)))).))))).))))...))))))) ( -43.20) >DroEre_CAF1 49810 120 + 1 GCCCAUCGCAUGGGUCAGACAAAAAAGGUGAUGAUCUACCGAUUUGUGACCCACAACUCGGUCGAGGAGCGCAUGAUGCAGGUUGCCAAGCACAAGAUGAUGCUCACUCAUCUUGUGGUG ((.((((((.((((((..(((((...((((......))))..)))))))))))...(((....)))..))).)))..))...........((((((((((.......))))))))))... ( -39.90) >DroYak_CAF1 79777 120 + 1 GCUCAUCGCAUGGGUCAGACAAAGAAGGUGAUGAUCUACCGAUUUGUGACCCACAACUCCGUGGAGGAGCGCAUCAUGCAGGUUGUCAAACACAAGAUGAUGCUCACUCAUCUUGUGGUG ((((......((((((..(((((...((((......))))..)))))))))))...(((....)))))))((.....))...........((((((((((.......))))))))))... ( -42.70) >consensus GCCCAUCGCAUGGGACAGAAAAAGAAGGUAAUGAUCUACCGAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCACAAGAUGAUGCUCACCCAUCUUGUGGUG ((((((...))))).)..........((((((..((((((((.(((((...))))).)))))))).....((.....))..))))))...(((((((((.........)))))))))... (-38.10 = -37.70 + -0.40)


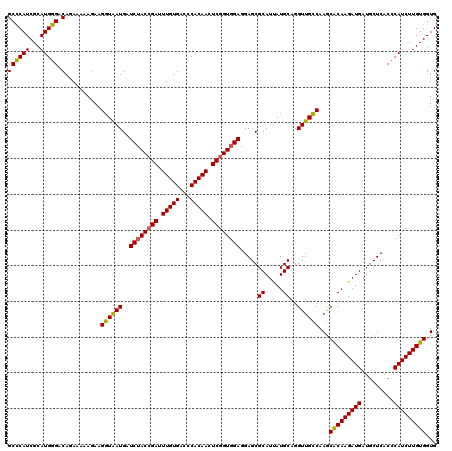
| Location | 19,414,632 – 19,414,752 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -36.22 |
| Energy contribution | -35.98 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19414632 120 + 23771897 GAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCACAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCAUGACAACAAAUUUCUCAAAGG ((.(((((...))))).))(((((..(((((((((.((...(((....))).)).)))).)))))..)))))((((.(((((((((....))))).)))).))))............... ( -41.00) >DroSec_CAF1 53404 120 + 1 GAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCGCAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCACGGCAACAAAUUUCUCAAAGG (((((((..(((((((...(((((..(((((((((.(((..(((....)))))).)))).)))))..))))))))))(((((((((....))))).))))))..)))))))......... ( -46.80) >DroSim_CAF1 54489 120 + 1 GAUUUGUGACUCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCGCAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCACGGCAACACAUUUCUCAAAGG ((..((((...........(((((..(((((((((.(((..(((....)))))).)))).)))))..)))))((((.(((((((((....))))).)))).))))))))..))....... ( -47.30) >DroEre_CAF1 49850 120 + 1 GAUUUGUGACCCACAACUCGGUCGAGGAGCGCAUGAUGCAGGUUGCCAAGCACAAGAUGAUGCUCACUCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCAAGGAAGUAAAUUUCACAAAGG ...((((.(((((...(((((.......((((((...((.....))....((((((((((.......)))))))))))))))))))))..))))).))))((((.....))))....... ( -42.51) >DroYak_CAF1 79817 120 + 1 GAUUUGUGACCCACAACUCCGUGGAGGAGCGCAUCAUGCAGGUUGUCAAACACAAGAUGAUGCUCACUCAUCUUGUGGUGCGCCCGGGAAUGGGCGGCAACAACAACAAUUUCACAAAGG ..(((((((.......((((.....)))).((.....))..((((((...((((((((((.......))))))))))....(((((....)))))))))))..........))))))).. ( -43.90) >consensus GAUUUGUGACCCACAACUCGGUGGAGGAGCGCAUUAUGCAGGUUGCCAAGCACAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCAAGGCAACAAAUUUCUCAAAGG ..((((.((.((((((...(((((..(((((((((.((...(((....))).)).)))).)))))..)))))))))))((((((((....))))).)))............)).)))).. (-36.22 = -35.98 + -0.24)

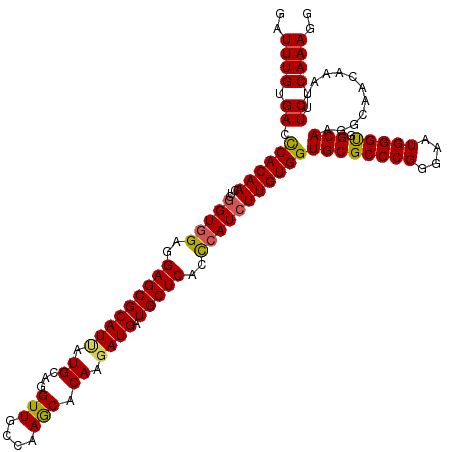
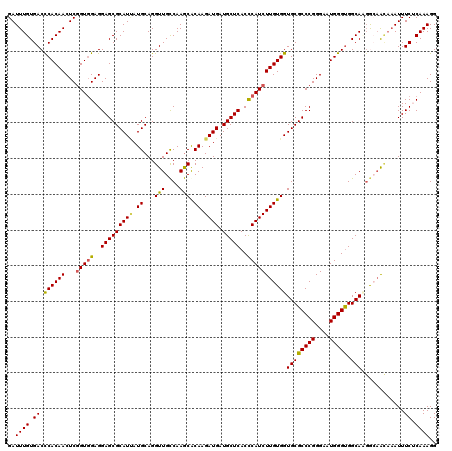
| Location | 19,414,632 – 19,414,752 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -31.94 |
| Energy contribution | -31.58 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19414632 120 - 23771897 CCUUUGAGAAAUUUGUUGUCAUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGUGCUUGGCAACCUGCAUAAUGCGCUCCUCCACCGAGUUGUGGGUCACAAAUC ....(((.((.....)).)))((..((((((..(((((((((..(((((((((.......)))))))))((..(....)..))....))))).......))).)..))))))..)).... ( -35.31) >DroSec_CAF1 53404 120 - 1 CCUUUGAGAAAUUUGUUGCCGUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGCGCUUGGCAACCUGCAUAAUGCGCUCCUCCACCGAGUUGUGGGUCACAAAUC ..........((((((.(((((((..(((......))).))))((((((.((((.((((..(((..((((...(....).))))..))).)))).)))).)...)))))))).)))))). ( -38.00) >DroSim_CAF1 54489 120 - 1 CCUUUGAGAAAUGUGUUGCCGUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGCGCUUGGCAACCUGCAUAAUGCGCUCCUCCACCGAGUUGUGAGUCACAAAUC ..((((.((.(((((((((((.((((((((((....((....))....)))))))).(((......))))).)))))))..))))....(((..(((....)))..)))..)).)))).. ( -38.70) >DroEre_CAF1 49850 120 - 1 CCUUUGUGAAAUUUACUUCCUUGCCACCCAUUCCCGGGCGCACCACAAGAUGAGUGAGCAUCAUCUUGUGCUUGGCAACCUGCAUCAUGCGCUCCUCGACCGAGUUGUGGGUCACAAAUC ..(((((((......(......)...(((((....(((((((..(((((((((.......)))))))))((..(....)..))....)))))))(((....)))..)))))))))))).. ( -39.00) >DroYak_CAF1 79817 120 - 1 CCUUUGUGAAAUUGUUGUUGUUGCCGCCCAUUCCCGGGCGCACCACAAGAUGAGUGAGCAUCAUCUUGUGUUUGACAACCUGCAUGAUGCGCUCCUCCACGGAGUUGUGGGUCACAAAUC ..(((((.....(((.((((((((.((((......))))))..((((((((((.......))))))))))...))))))..))).(((.(((..(((....)))..))).)))))))).. ( -44.90) >consensus CCUUUGAGAAAUUUGUUGCCGUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGUGCUUGGCAACCUGCAUAAUGCGCUCCUCCACCGAGUUGUGGGUCACAAAUC .....................((..((((((....(((((((...((((((((.......)))))))).((..(....)..))....)))))))(((....)))..))))))..)).... (-31.94 = -31.58 + -0.36)

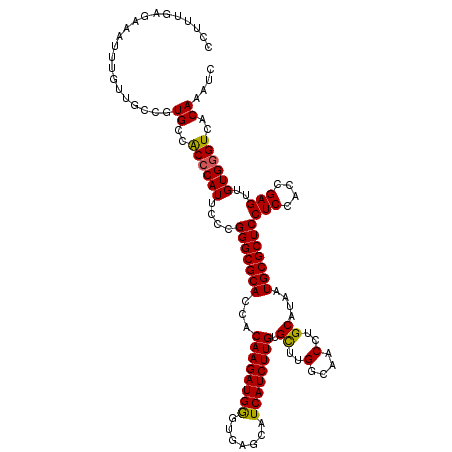

| Location | 19,414,672 – 19,414,792 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -42.34 |
| Consensus MFE | -36.20 |
| Energy contribution | -35.80 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19414672 120 + 23771897 GGUUGCCAAGCACAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCAUGACAACAAAUUUCUCAAAGGACGAGCUCGAAGACAUCCUUCGCUUUGGCACCGAGGAUCU (((.(((((((....(((.((((.(((((((((((.((....)))))).))))))))))).)..........(((.......))).))((((.....)))))).))))))))........ ( -41.00) >DroSec_CAF1 53444 120 + 1 GGUUGCCAAGCGCAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCACGGCAACAAAUUUCUCAAAGGACGAGCUCGAAGACAUCCUUCGCUUUGGCACCGAGGAUCU .((((((...(((((((((.........)))))))))(((((((((....))))).))))))))))....(((((...((.((((..(((((.....))))).))))...)))))))... ( -45.10) >DroSim_CAF1 54529 120 + 1 GGUUGCCAAGCGCAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCACGGCAACACAUUUCUCAAAGGACGAGCUCGAAGACAUCCUUCGCUUUGGCACCGAGGAUCU .((((((...(((((((((.........)))))))))(((((((((....))))).))))))))))....(((((...((.((((..(((((.....))))).))))...)))))))... ( -45.10) >DroEre_CAF1 49890 120 + 1 GGUUGCCAAGCACAAGAUGAUGCUCACUCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCAAGGAAGUAAAUUUCACAAAGGACGAGCUCGCUGACAUCCUUCGCUUUGGCACCGAGGAUCU (((.((((((((((((((((.......))))))))))...((((((....))))))(((((((.((............((.((....)))).)).))))).)))))))))))........ ( -41.22) >DroYak_CAF1 79857 120 + 1 GGUUGUCAAACACAAGAUGAUGCUCACUCAUCUUGUGGUGCGCCCGGGAAUGGGCGGCAACAACAACAAUUUCACAAAGGACGAGCUCGAAAACAUCCUUCGCUUUGGCACCGAGGAUCU ((((.((....(((((((((.......)))))))))((((((((((....)))))(....).............(((((..((((...((.....)).)))))))))))))))).)))). ( -39.30) >consensus GGUUGCCAAGCACAAGAUGAUGCUCACCCAUCUUGUGGUGCGCCCGGGAAUGGGUGGCAAGGCAACAAAUUUCUCAAAGGACGAGCUCGAAGACAUCCUUCGCUUUGGCACCGAGGAUCU (((.(((((((((((((((.........)))))))))(((((((((....))))).))))................(((((((....))......)))))...)))))))))........ (-36.20 = -35.80 + -0.40)

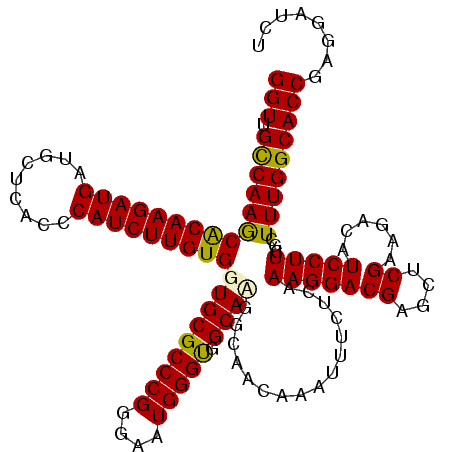

| Location | 19,414,672 – 19,414,792 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -42.06 |
| Consensus MFE | -35.42 |
| Energy contribution | -35.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19414672 120 - 23771897 AGAUCCUCGGUGCCAAAGCGAAGGAUGUCUUCGAGCUCGUCCUUUGAGAAAUUUGUUGUCAUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGUGCUUGGCAACC ........((((((((.(((.((((((((((.(((......))).))))...........((((((((((((....((....))....)))))))).)))))))))).)))))))).))) ( -41.20) >DroSec_CAF1 53444 120 - 1 AGAUCCUCGGUGCCAAAGCGAAGGAUGUCUUCGAGCUCGUCCUUUGAGAAAUUUGUUGCCGUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGCGCUUGGCAACC ........((((((((.(((.((((((((((.(((......))).))))...........((((((((((((....((....))....)))))))).)))))))))).)))))))).))) ( -43.10) >DroSim_CAF1 54529 120 - 1 AGAUCCUCGGUGCCAAAGCGAAGGAUGUCUUCGAGCUCGUCCUUUGAGAAAUGUGUUGCCGUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGCGCUUGGCAACC ........((((((((.(((.((((((((((.(((......))).))))...........((((((((((((....((....))....)))))))).)))))))))).)))))))).))) ( -43.10) >DroEre_CAF1 49890 120 - 1 AGAUCCUCGGUGCCAAAGCGAAGGAUGUCAGCGAGCUCGUCCUUUGUGAAAUUUACUUCCUUGCCACCCAUUCCCGGGCGCACCACAAGAUGAGUGAGCAUCAUCUUGUGCUUGGCAACC ........((((((((.((((((((((..((....))))))))))))..............(((..(((......))).))).((((((((((.......)))))))))).))))).))) ( -41.50) >DroYak_CAF1 79857 120 - 1 AGAUCCUCGGUGCCAAAGCGAAGGAUGUUUUCGAGCUCGUCCUUUGUGAAAUUGUUGUUGUUGCCGCCCAUUCCCGGGCGCACCACAAGAUGAGUGAGCAUCAUCUUGUGUUUGACAACC .................((((((((((..........))))))))))......((((((..(((.((((......))))))).((((((((((.......))))))))))...)))))). ( -41.40) >consensus AGAUCCUCGGUGCCAAAGCGAAGGAUGUCUUCGAGCUCGUCCUUUGAGAAAUUUGUUGCCGUGCCACCCAUUCCCGGGCGCACCACAAGAUGGGUGAGCAUCAUCUUGUGCUUGGCAACC ........((((((((.(((.((((((((((.(((......))).))))............(((((((((((....((....))....)))))))).))).)))))).)))))))).))) (-35.42 = -35.46 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:21 2006