
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,407,969 – 19,408,083 |
| Length | 114 |
| Max. P | 0.686917 |

| Location | 19,407,969 – 19,408,065 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -15.10 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19407969 96 - 23771897 GCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCUCCUCUUUCCUCAAGCUC--------- ((..((((((....)))))).....((..(((((......))))).....(((((...)))))..)).........................))..--------- ( -15.80) >DroSec_CAF1 46831 96 - 1 GCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACCGAAUUUCCUCCUCUUUCCCCAAGCCC--------- ((..((((((....))))))...(((((.(((((......)))))....)))))...........)).............................--------- ( -13.50) >DroSim_CAF1 46994 96 - 1 GCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCACCUCUUUCCCCAAGCUC--------- ((..((((((....)))))).....((..(((((......))))).....(((((...)))))..)).........................))..--------- ( -15.80) >DroEre_CAF1 43298 96 - 1 CCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUCCCUCCUCCUUCCCCAACUUC--------- ....((((((....)))))).....((..(((((......))))).....(((((...)))))..)).............................--------- ( -13.60) >DroYak_CAF1 72955 96 - 1 CCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCUCCUCUUUCCCCAAGCUC--------- ....((((((....)))))).....((..(((((......))))).....(((((...)))))..)).............................--------- ( -13.60) >DroAna_CAF1 45675 105 - 1 CCAAAGUCAACCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACGAGACUGCCACCUCUCCCCACACUCUCCGCAAUUUC ....((((((....))))))...(((((.(((((......)))))....))))).....((((((....).)))))............................. ( -18.30) >consensus CCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCUCCUCUUUCCCCAAGCUC_________ ....((((((....)))))).....((..(((((......))))).....((((.....))))..))...................................... (-12.69 = -12.47 + -0.22)

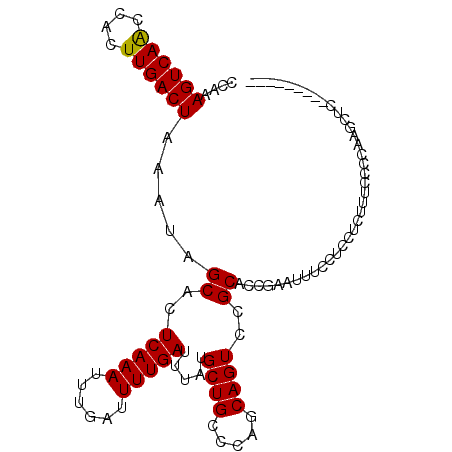

| Location | 19,407,993 – 19,408,083 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -14.18 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19407993 90 - 23771897 ----CAUC-UAU------CCCCACU---CCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGA ----....-...------.......---..((((..((((((....))))))...(((((.(((((......)))))....)))))....)))).......... ( -14.60) >DroSec_CAF1 46855 90 - 1 ----CAUC-CAU------UCCCACU---CCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACCGA ----....-...------.......---...(((..((((((....))))))...(((((.(((((......)))))....)))))...........))).... ( -14.10) >DroSim_CAF1 47018 94 - 1 ACAAGAUU-CAU------CCCCACU---CCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGA ........-...------.......---..((((..((((((....))))))...(((((.(((((......)))))....)))))....)))).......... ( -14.60) >DroEre_CAF1 43322 85 - 1 ----CA-----U------CACCAU----CCUUCCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGA ----..-----.------......----........((((((....)))))).....((..(((((......))))).....(((((...)))))..))..... ( -13.60) >DroYak_CAF1 72979 91 - 1 ----CAUCCCAU------CACCAUU---CCUUCCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGA ----........------.......---........((((((....)))))).....((..(((((......))))).....(((((...)))))..))..... ( -13.60) >DroAna_CAF1 45708 100 - 1 ----CAACCGCCCAACCGAACCAUUCUCCCCCCCAAAGUCAACCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACGAG ----.....((.........................((((((....))))))...(((((.(((((......)))))....)))))...........))..... ( -14.60) >consensus ____CAUC_CAU______CACCACU___CCUUCCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGA ....................................((((((....)))))).....((..(((((......))))).....((((.....))))..))..... (-12.69 = -12.47 + -0.22)

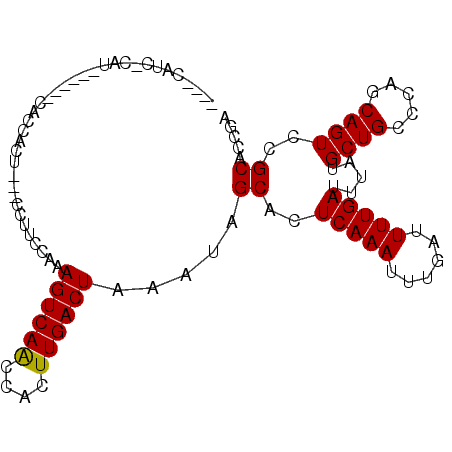
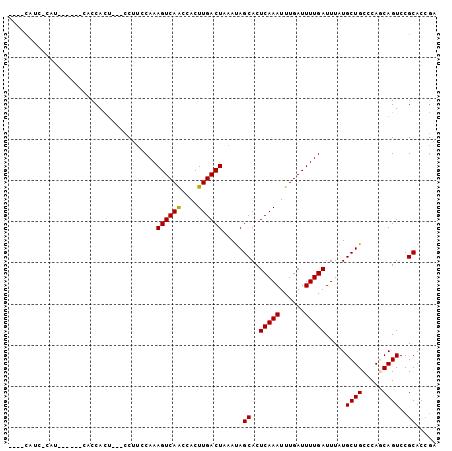
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:08 2006