| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,407,771 – 19,407,874 |
| Length | 103 |
| Max. P | 0.956115 |

| Location | 19,407,771 – 19,407,874 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19407771 103 + 23771897 -----AUAUUCAAAUA---UAGCAGAUAUAGAUAUUUCUUUGAGUGUU--AGUUUCACUCCGUUUCCGUUACACGCCGAUGCGCGUGUAAGGAGCAUGUAAAUAUGAAAGGCA -----...((((..((---((((..................(((((..--.....)))))....(((.((((((((......))))))))))))).))))....))))..... ( -24.00) >DroSec_CAF1 46640 100 + 1 -----UUAUGGAAAUA---UAGAA--UAUUGCU-UUUCUCUGAGUGUA--CGUUUCACUCCGUUUCCGUUACACGCCGAUGCGCGUGUAAGGAGCAUGUAAAUAUGAAAGGCA -----...........---.....--...((((-((((...(((((..--.....)))))(((((((.((((((((......)))))))))))).))).......)))))))) ( -29.70) >DroSim_CAF1 46803 100 + 1 -----UUAUGCAAAUA---UAGAA--UAAUGCU-UUUCUCUGAGUGUA--CGUUUCACUCCGUUUCCGUUACACGCCGAUGCGCGUGUAAGGAGCAUGUAAAUAUGAAAGGCA -----...........---.....--...((((-((((...(((((..--.....)))))(((((((.((((((((......)))))))))))).))).......)))))))) ( -29.80) >DroEre_CAF1 43114 95 + 1 -----AAAUUCCAACA---AACAC-------AA-UUUCGCUGCGUGUA--AAUAGCGCUCCGUCUCCGUUACACGCCGAUGCGCGUGUAAGGAGCAUGUAAAUAUGAAAGGCA -----...........---.....-------..-....(((((((...--....))))..(((((((.((((((((......)))))))))))).)))...........))). ( -26.20) >DroYak_CAF1 72772 95 + 1 -----AAAUCCCAAUA---AACAC-------AU-UUUCGCUGCGUGUA--AAUGGCACUCCUUCUCUGUUACACGCCGAUGCGCGUGUAAGGAGCAUGUAAAUAUGAAAGGCA -----...........---.....-------..-....(((.(((((.--....((((((((.......(((((((......))))))))))))..)))..)))))...))). ( -22.21) >DroAna_CAF1 45456 107 + 1 AUGAAAACUUCCUACAUCUUAGUC--UA---AU-UUUUUUUCAAUGCCAGAAUUCUACUCAAUUUCCGUUACACGCCAAUGCGCGUGUAAGGAGCAUGUAAAUAUGAAAGGCA .((((((......((......)).--..---..-...)))))).((((...............((((.((((((((......))))))))))))((((....))))...)))) ( -21.74) >consensus _____AAAUUCAAAUA___UAGAA__UA___AU_UUUCUCUGAGUGUA__AAUUUCACUCCGUUUCCGUUACACGCCGAUGCGCGUGUAAGGAGCAUGUAAAUAUGAAAGGCA ..................................((((...(((((.........)))))(((((((.((((((((......)))))))))))).))).......)))).... (-17.65 = -17.77 + 0.12)

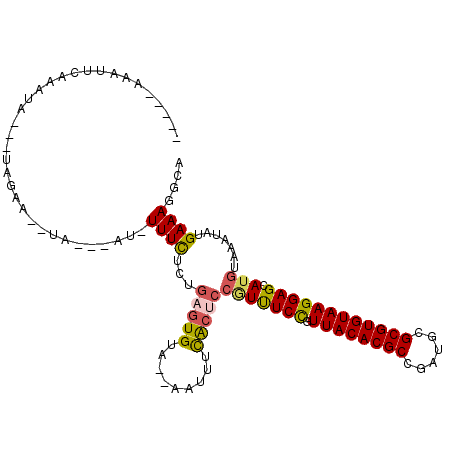

| Location | 19,407,771 – 19,407,874 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -17.39 |
| Energy contribution | -17.33 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19407771 103 - 23771897 UGCCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAAACGGAGUGAAACU--AACACUCAAAGAAAUAUCUAUAUCUGCUA---UAUUUGAAUAU----- ........(((((((.(((.(((((((((((......)))))))).)))....(((((.....--..)))))......................---))).)))))))----- ( -23.40) >DroSec_CAF1 46640 100 - 1 UGCCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAAACGGAGUGAAACG--UACACUCAGAGAAA-AGCAAUA--UUCUA---UAUUUCCAUAA----- (((.((((............(((((((((((......)))))))).)))....(((((.....--..)))))...))))-.)))...--.....---...........----- ( -27.00) >DroSim_CAF1 46803 100 - 1 UGCCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAAACGGAGUGAAACG--UACACUCAGAGAAA-AGCAUUA--UUCUA---UAUUUGCAUAA----- (((.((((............(((((((((((......)))))))).)))....(((((.....--..)))))...))))-.)))...--.....---...........----- ( -26.80) >DroEre_CAF1 43114 95 - 1 UGCCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAGACGGAGCGCUAUU--UACACGCAGCGAAA-UU-------GUGUU---UGUUGGAAUUU----- ....(((((((..((((.(((((((((((((......)))))))).(....))))))(((...--.......)))....-.)-------)))..---)).)))))...----- ( -28.70) >DroYak_CAF1 72772 95 - 1 UGCCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACAGAGAAGGAGUGCCAUU--UACACGCAGCGAAA-AU-------GUGUU---UAUUGGGAUUU----- ..................(((((((((((((......))))))).......)))))).(((..--((.(((((......-.)-------)))).---)).))).....----- ( -24.21) >DroAna_CAF1 45456 107 - 1 UGCCUUUCAUAUUUACAUGCUCCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGUAGAAUUCUGGCAUUGAAAAAAA-AU---UA--GACUAAGAUGUAGGAAGUUUUCAU ((((.(((.((((((.((..(((((((((((......)))))))).))).)))))))))))....)))).((((((...-.(---(.--.((......))..))..)))))). ( -27.90) >consensus UGCCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAAACGGAGUGAAACU__UACACUCAGAGAAA_AU___UA__GUCUA___UAUUGGAAUAU_____ .................((((((((((((((......)))))))).(....)))))))....................................................... (-17.39 = -17.33 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:06 2006