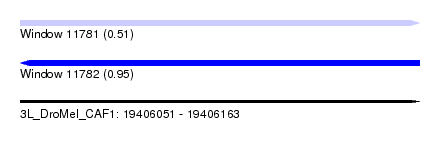
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,406,051 – 19,406,163 |
| Length | 112 |
| Max. P | 0.952589 |

| Location | 19,406,051 – 19,406,163 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -13.07 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
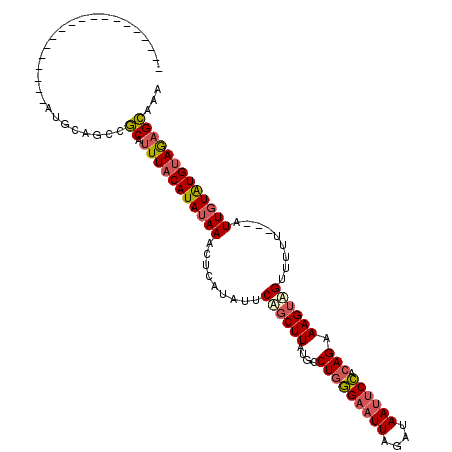
>3L_DroMel_CAF1 19406051 112 + 23771897 UUUGCUCUGCACACAAU---AAAAACUACUUUCUGUGGAAUUAUCUAAUUCCCAGGCAUAAGCAGAAUAUGAGUUUAUAUGUAAAUGCGGCUGCAUAUCCAAGCCGCUCAUAUCC ((((((.(((.......---(((......)))(((.((((((....))))))))))))..))))))(((((((((((....)))))((((((.........)))))))))))).. ( -28.20) >DroSec_CAF1 44967 92 + 1 UUUGCUCUACACACAAU---AAAAACUACUUUCUGUGGAAUUAUCUAAU-CCCAGGCAUAAGCUGAAUACGAGUUUAUAUGUAAAUGCGGUCGCAU------------------- .((((..((((......---...........((((.(((.........)-)))))).(((((((.......))))))).))))...))))......------------------- ( -13.40) >DroSim_CAF1 45129 93 + 1 UUUGCUCUACAUACAAU---AAAAACUACUUUCUGUGGAAUUAUCUAAUUCCCAGGCAUAAGCUGAAUACGAGUUUAUAUGUAAAUGCGGCUGCAU------------------- ..(((.(..((((((..---...........((((.((((((....)))))))))).(((((((.......))))))).))))..))..)..))).------------------- ( -18.50) >DroYak_CAF1 71029 96 + 1 AUCACAUUACAUAAAAUGUUUAAAUCCACUUUCUUUAGAUUUAUCUAAUUCUGAGGUAUAAGAGGAAUGUGAGUUUAUAUGUAAAUGUGGCUGCAU------------------- .((((((((((((((((.......(((.(((.(((((((..........)))))))...))).)))......)))).))))).)))))))......------------------- ( -21.52) >consensus UUUGCUCUACACACAAU___AAAAACUACUUUCUGUGGAAUUAUCUAAUUCCCAGGCAUAAGCUGAAUACGAGUUUAUAUGUAAAUGCGGCUGCAU___________________ .((((..((((....................((((.((((((....)))))))))).(((((((.......))))))).))))...))))......................... (-13.07 = -12.82 + -0.25)

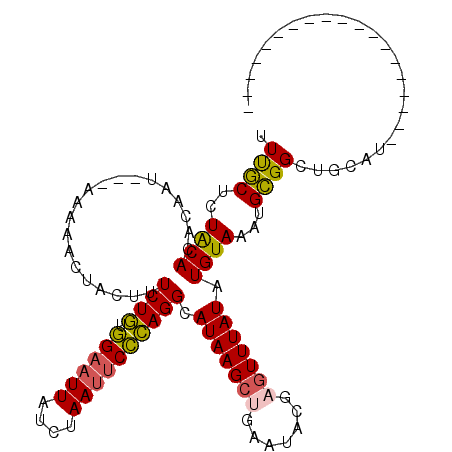
| Location | 19,406,051 – 19,406,163 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -16.43 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19406051 112 - 23771897 GGAUAUGAGCGGCUUGGAUAUGCAGCCGCAUUUACAUAUAAACUCAUAUUCUGCUUAUGCCUGGGAAUUAGAUAAUUCCACAGAAAGUAGUUUUU---AUUGUGUGCAGAGCAAA ((((((((((((((.(......))))))).((((....)))).))))))))(((((.((((((((((((....)))))).)))((((....))))---.......)))))))).. ( -31.40) >DroSec_CAF1 44967 92 - 1 -------------------AUGCGACCGCAUUUACAUAUAAACUCGUAUUCAGCUUAUGCCUGGG-AUUAGAUAAUUCCACAGAAAGUAGUUUUU---AUUGUGUGUAGAGCAAA -------------------........((.(((((((((((...........((....))(((((-(.........))).)))............---.)))))))))))))... ( -19.20) >DroSim_CAF1 45129 93 - 1 -------------------AUGCAGCCGCAUUUACAUAUAAACUCGUAUUCAGCUUAUGCCUGGGAAUUAGAUAAUUCCACAGAAAGUAGUUUUU---AUUGUAUGUAGAGCAAA -------------------........((.(((((((((((...........((....))(((((((((....)))))).)))............---.)))))))))))))... ( -22.30) >DroYak_CAF1 71029 96 - 1 -------------------AUGCAGCCACAUUUACAUAUAAACUCACAUUCCUCUUAUACCUCAGAAUUAGAUAAAUCUAAAGAAAGUGGAUUUAAACAUUUUAUGUAAUGUGAU -------------------.......((((((.(((((((((..(((.(((.(((........))).(((((....))))).))).)))..)))).......))))))))))).. ( -16.71) >consensus ___________________AUGCAGCCGCAUUUACAUAUAAACUCAUAUUCAGCUUAUGCCUGGGAAUUAGAUAAUUCCACAGAAAGUAGUUUUU___AUUGUAUGUAGAGCAAA ...........................((.(((((((((((.........((((((....(((((((((....)))))).))).)))))).........)))))))))))))... (-16.43 = -17.37 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:04 2006