
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,402,811 – 19,402,943 |
| Length | 132 |
| Max. P | 0.601290 |

| Location | 19,402,811 – 19,402,911 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.96 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.11 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
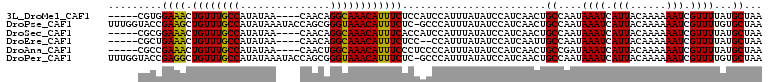
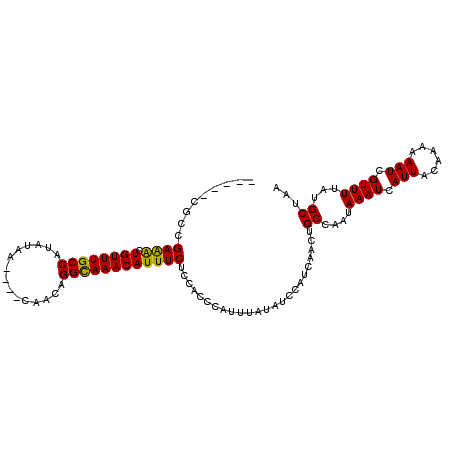
>3L_DroMel_CAF1 19402811 100 - 23771897 -----CGUGGAAACUGUUUGCCAUAUAA----CAACAGGCAAACAUUUCUCCAUCCAUUUAUAUCCAUCAACUGCCAAUAAAUCAUUACAAAAAAUCGUUUUAUGCUAA -----.(((((((.((((((((......----.....)))))))).)).)))))...................((....((((.(((......))).))))...))... ( -15.70) >DroPse_CAF1 10556 108 - 1 UUUGGUACCGAAGCUGUUUGCCAUAUAAAUACCAGCGGGUAAACAUUUCUC-GCCCAUUUAUAUCCAUCAACUGCCAAUAAAUCAUUACAAAAAAUCGUUUUGUGCUAA .((((((.....((.....)).((((((((....(((((.........)))-))..))))))))........))))))........(((((((.....))))))).... ( -17.70) >DroSec_CAF1 42038 100 - 1 -----CGCGGAAACUGUUUGCCAUAUAA----CAACAGGCAAACAUUUCACCAUCCAUUUAUAUCCAUCAACUGCCAAUAAAUCAUUACAAAAAAUCGUUUUAUGCUAA -----.(((((((.((((((((......----.....)))))))))))).......((((((...((.....))...))))))....................)))... ( -14.60) >DroEre_CAF1 36904 98 - 1 -----CGCUGAAACUGUUUGCCAUAUAA----CAACAGGCAAACAUUUCUCC--CCAUUUAUAUCCAUCAAUUGCCAAUAAAUCAUUACAAAAAAUCGUUUUAUGCUAA -----.((((((((((((((((......----.....)))))))).......--..((((((...((.....))...))))))..............)))))).))... ( -16.50) >DroAna_CAF1 40985 100 - 1 -----CGCCGAAACUGUUUGCCAUAUAA----CAACUGGCAAACAUUUCCCUCCCCAUUUAUAUCCAUCAACUGCCGAUAAAUCAUUACAAAAAAUCGUUUUAUGCUAA -----.((.((((((((((((((.....----....)))))))))...........((((((...((.....))...))))))..............)))))..))... ( -15.60) >DroPer_CAF1 10570 108 - 1 UUUGGUACCGAGGCUGUUUGCCAUAUAAAUACCAGCGGGUAAACAUUUCUC-GCCCAUUUAUAUCCAUCAACUGCCAAUAAAUCAUUACAAAAAAUCGUUUUGUGCUAA .((((((....(((.....)))((((((((....(((((.........)))-))..))))))))........))))))........(((((((.....))))))).... ( -21.90) >consensus _____CGCCGAAACUGUUUGCCAUAUAA____CAACAGGCAAACAUUUCUCCACCCAUUUAUAUCCAUCAACUGCCAAUAAAUCAUUACAAAAAAUCGUUUUAUGCUAA .........((((.((((((((...............))))))))))))........................((....((((.(((......))).))))...))... (-12.69 = -12.11 + -0.58)
| Location | 19,402,842 – 19,402,943 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
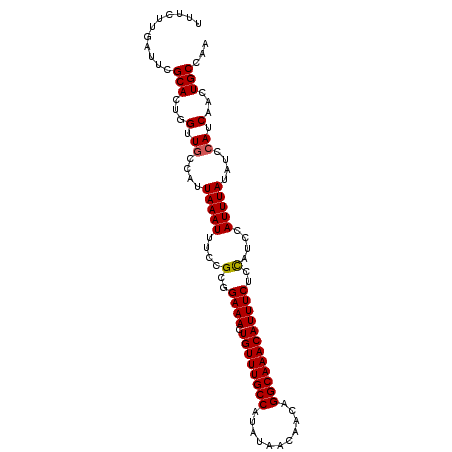
>3L_DroMel_CAF1 19402842 101 - 23771897 -UUCUUGGUUCGCACUGGUUGCCAUUAAAUUUCCGUGGAAACUGUUUGCCAUAUAACAACAGGCAAACAUUUCUCCAUCCAUUUAUAUCCAUCAACUGCCAA -....((((......(((.......(((((....(((((((.((((((((...........)))))))).)).)))))..)))))...)))......)))). ( -21.70) >DroSec_CAF1 42069 102 - 1 UUUCUUGAUUCGCACUGGUUGCCAUUAAAUUUCCGCGGAAACUGUUUGCCAUAUAACAACAGGCAAACAUUUCACCAUCCAUUUAUAUCCAUCAACUGCCAA ....(((((.(((...((((.......))))...)))((((.((((((((...........)))))))))))).................)))))....... ( -18.40) >DroSim_CAF1 41966 102 - 1 UUUCUUGAUUCGCACUGGUUGCCAUUAAAUUUCCGCGGAAACUGUUUGCCAUAUAACAACAGGCAAACAUUUCUCCAUCCAUUUAUAUCCAUCAACUGCCAA ....(((((.(((...((((.......))))...)))((((.((((((((...........)))))))))))).................)))))....... ( -17.90) >DroEre_CAF1 36935 100 - 1 UUGCUUGAUCCGCACUGGUUGCCAUUAAAUUUCCGCUGAAACUGUUUGCCAUAUAACAACAGGCAAACAUUUCUCC--CCAUUUAUAUCCAUCAAUUGCCAA ..(((((((..(((.....)))...............((((.((((((((...........))))))))))))...--............)))))..))... ( -16.40) >DroYak_CAF1 67756 102 - 1 UUGCUUGAUUUGCACGGGCUGCCAUUAAAUUUCCGCGGAAACUGUUUGCCAUAUAACAACAGGCAAACAUUUCUCCCCCCAUUUAUAUCCAUCAACUGCCAA .(((.......)))..(((.((............))(((((.((((((((...........))))))))))))).......................))).. ( -19.40) >DroAna_CAF1 41016 102 - 1 UUUCUUGGCUCGCAGUUGUUCCCAUUAAAUUUACGCCGAAACUGUUUGCCAUAUAACAACUGGCAAACAUUUCCCUCCCCAUUUAUAUCCAUCAACUGCCGA ...........(((((((.......(((((.......((((.(((((((((.........))))))))))))).......))))).......)))))))... ( -24.78) >consensus UUUCUUGAUUCGCACUGGUUGCCAUUAAAUUUCCGCGGAAACUGUUUGCCAUAUAACAACAGGCAAACAUUUCUCCAUCCAUUUAUAUCCAUCAACUGCCAA ...........(((...(.((....(((((....(..((((.((((((((...........))))))))))))..)....)))))....)).)...)))... (-15.65 = -15.68 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:03 2006