| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,402,059 – 19,402,166 |
| Length | 107 |
| Max. P | 0.580458 |

| Location | 19,402,059 – 19,402,166 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -19.63 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
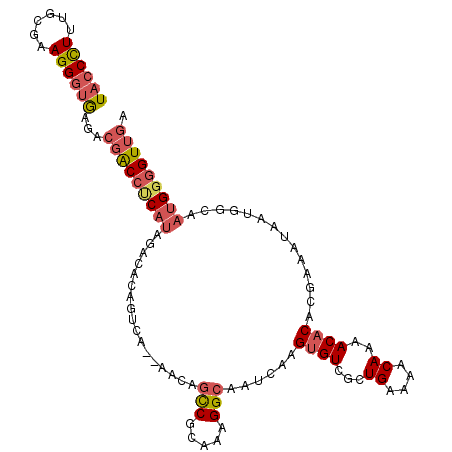
>3L_DroMel_CAF1 19402059 107 - 23771897 UACCCUUUGUGAAGGGUGUGACGACCUCAUAGACACACUCA--AACAGCCGCAAAGGCAAUCAAGUGUCGCUGAAAACAAAACACACAACAUAAUGAGAAUGGGGUUGU (((((((....)))))))..((((((((((.(((((.....--....(((.....)))......))))).................((......))...)))))))))) ( -29.06) >DroSec_CAF1 41285 107 - 1 UACCUUUUCUACAGGGUGUAACGACCUCAUAGACACAGUCA--AACAGCCGCAAAGGCAAUCAAGUGUCCCUGAAAACAAAACACACGACAUAAUGGCAAUGGGGUUGA ((((((......))))))...(((((((((.(.((..(((.--....(((.....)))......((((...((....))..))))..)))....)).).))))))))). ( -27.00) >DroSim_CAF1 41193 107 - 1 UACCCUUUCUGCAGGGUGUGACGACCUCAUAGACACAGUCA--AACAGCCGCAAAGGCAAUCAAGUGUCGCUGAAAACAAAACACACGACAUAAUGGCAAUGGAGUUGA ((((((......))))))...((((..(((.(((...))).--....(((((....))......((((((.((...........))))))))...))).)))..)))). ( -27.50) >DroEre_CAF1 36186 107 - 1 UACCCUUUGCGAAGGGUAAGCCGACCGCAUAGAUACAGUCA--AACUGCCGCAAAGGCAAUCAAGUGUCGCUGAAAACAAAACACACGAAAUAAUGGCAAUGGGGUUGA (((((((....)))))))...(((((.(((.(((((((...--..))(((.....)))......)))))(((.......................))).))).))))). ( -29.60) >DroYak_CAF1 66957 107 - 1 UACCUUUUGCGAAGGGUAAGCCGACCUCAUAGACACAUUCA--AACAGCCGCAAAGGCAAUCAAGGGUCGCUGAAAACAAAACACACGAAAUAAUGGCAAUGGGGUUGA (((((((....)))))))...(((((((((.(...((((..--....(((.....))).........(((.((...........)))))...)))).).))))))))). ( -25.20) >DroAna_CAF1 40161 106 - 1 UACCCUAAUC-AAGGUAGAGAA-GCCCCACAGACAGACCCACUCACACUCGCACAGGCAAUCAAGUGUCGUUGAAAACAAAACACACGAAA-AAUAUAAAAGGGGUUGG ((((......-..))))....(-(((((......................((....))......((((.(((........)))))))....-.........)))))).. ( -20.10) >consensus UACCCUUUGCGAAGGGUGAGACGACCUCAUAGACACAGUCA__AACAGCCGCAAAGGCAAUCAAGUGUCGCUGAAAACAAAACACACGAAAUAAUGGCAAUGGGGUUGA ((((((......))))))...(((((((((.................(((.....)))......((((...((....))..))))..............))))))))). (-19.63 = -20.47 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:01 2006