
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,398,364 – 19,398,548 |
| Length | 184 |
| Max. P | 0.996731 |

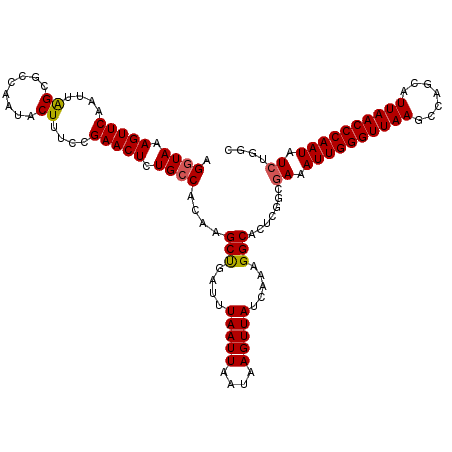
| Location | 19,398,364 – 19,398,482 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19398364 118 + 23771897 AGGUAAAGUUCAAUUAGCGCCAAUACUUUCCGAACUCUGCCACAAGCUGAUUUAAUUAAUAAGUUAUAAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGC ..................((((.......((((....((((......(((((((.....))))))).....)))).)))).((.((((((((((.......)))))))))).)))))) ( -28.40) >DroSec_CAF1 37577 118 + 1 AGGUAAAGUUCAAUUGGCGCCAAUACUUUCCGAACUCUGCCACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGAUGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGC .((.(((((...((((....))))))))))).......((((...(((....(((((....))))).....))).......((.((((((((((.......)))))))))).)))))) ( -28.50) >DroSim_CAF1 37466 118 + 1 AGGUAAAGUUCAAUUAGCGCCAAUACUUUCCGAACUCUGCCACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGC ..................((((.......((((....((((....(.(((((((.....))))))).)...)))).)))).((.((((((((((.......)))))))))).)))))) ( -28.40) >DroEre_CAF1 32519 118 + 1 GGGUAAAGUUCAAUUAGCGCCAAUACUUUCCGAACUCUGCCACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUCU .((((.(((((....((........))....))))).))))....(((....(((((....))))).....)))....((.((.((((((((((.......)))))))))).)).)). ( -27.50) >DroYak_CAF1 63191 118 + 1 AGCUAAAGUUCAAUUAGGGCCAAUACUUUCCGAACUCUGCCACAAGCCGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCAAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUUC ......(((((....((........))....))))).((((....(((....(((((....))))).....)))....))))..((((((((((.......))))))))))....... ( -26.70) >consensus AGGUAAAGUUCAAUUAGCGCCAAUACUUUCCGAACUCUGCCACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGC .((((.(((((....((........))....))))).))))....(((....(((((....))))).....))).......((.((((((((((.......)))))))))).)).... (-25.72 = -25.80 + 0.08)

| Location | 19,398,364 – 19,398,482 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19398364 118 - 23771897 GCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUUAUAACUUAUUAAUUAAAUCAGCUUGUGGCAGAGUUCGGAAAGUAUUGGCGCUAAUUGAACUUUACCU ((((((.((((((((((.(....))))))))))).))..(((((....((((............))))...)))))))))(((((((((..(((......)))..))))))))).... ( -32.30) >DroSec_CAF1 37577 118 - 1 GCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCAUCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUGGCAGAGUUCGGAAAGUAUUGGCGCCAAUUGAACUUUACCU ((((((.((((((((((.(....))))))))))).))..(((((...((((((.........))))))...)))))))))((((((((......((((....)))))))))))).... ( -32.70) >DroSim_CAF1 37466 118 - 1 GCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUGGCAGAGUUCGGAAAGUAUUGGCGCUAAUUGAACUUUACCU ((((((.((((((((((.(....))))))))))).))..(((((...((((((.........))))))...)))))))))(((((((((..(((......)))..))))))))).... ( -34.10) >DroEre_CAF1 32519 118 - 1 AGAAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUGGCAGAGUUCGGAAAGUAUUGGCGCUAAUUGAACUUUACCC ....((.((((((((((.(....))))))))))).))((((...((.((((((.........))))))...))...))))(((((((((..(((......)))..))))))))).... ( -31.00) >DroYak_CAF1 63191 118 - 1 GAAAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUUGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCGGCUUGUGGCAGAGUUCGGAAAGUAUUGGCCCUAAUUGAACUUUAGCU .((((..((((((((((((((.......((.((((((((((...(((((((((.........))))))..)))...)))))))))).))..))))))))))).)))....)))).... ( -34.50) >consensus GCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUGGCAGAGUUCGGAAAGUAUUGGCGCUAAUUGAACUUUACCU ....((.((((((((((.(....))))))))))).))((((...((.((((((.........))))))...))...))))(((((((((..(((......)))..))))))))).... (-26.86 = -27.34 + 0.48)


| Location | 19,398,404 – 19,398,515 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -25.86 |
| Energy contribution | -26.86 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19398404 111 + 23771897 CACAAGCUGAUUUAAUUAAUAAGUUAUAAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCACGGAUUAAAGCUCGU .((.((((...((((((.....((........((((.((((((.((((((((((.......)))))))))).))..)))))))).......)).....)))))))))).)) ( -32.76) >DroSec_CAF1 37617 111 + 1 CACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGAUGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCAAGGAUUAAAGCUCGU .((.((((...((((((.....((((.((..(((.(.....((.((((((((((.......)))))))))).)).))))..)))))).....(....))))))))))).)) ( -29.40) >DroSim_CAF1 37506 111 + 1 CACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCAAGGAUUAAAGCUCGU ....(((((...(((((....)))))......((((.((((((.((((((((((.......)))))))))).))..))))))))..))))).(....)............. ( -32.80) >DroEre_CAF1 32559 111 + 1 CACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUCUCGGUGUAACAGCUGCGCAAAGAUUAAAGCUCGU .....(((....(((((....))))).....)))...((((((.((((((((((.......)))))))))).)).(((..(((((.....)))))..))).....)).)). ( -27.20) >DroYak_CAF1 63231 111 + 1 CACAAGCCGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCAAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUUCCGUUGUAACAGCUGCGCAAAGAUUAAAGCUCGU .....(((....(((((....))))).....)))...((((...((((((((((.......))))))))))((((((.(((.((....)).))).))))))....)).)). ( -28.20) >consensus CACAAGCUGAUUUAAUUAAUAAGUUAUCAAAGGCACUCGGCGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCAAGGAUUAAAGCUCGU .((.((((...((((((.....((........((((.((((((.((((((((((.......)))))))))).))..)))))))).......)).....)))))))))).)) (-25.86 = -26.86 + 1.00)



| Location | 19,398,404 – 19,398,515 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19398404 111 - 23771897 ACGAGCUUUAAUCCGUGCGCAGCUGUUACACCGGCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUUAUAACUUAUUAAUUAAAUCAGCUUGUG ((((((((((((..(((...........(((((((..((.((((((((((.(....))))))))))).)))))).)))............)))..)))))...))))))). ( -30.30) >DroSec_CAF1 37617 111 - 1 ACGAGCUUUAAUCCUUGCGCAGCUGUUACACCGGCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCAUCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUG (((((((.........((((.((((......))))..((.((((((((((.(....))))))))))).)).....))))..((((((...)))))).......))))))). ( -30.50) >DroSim_CAF1 37506 111 - 1 ACGAGCUUUAAUCCUUGCGCAGCUGUUACACCGGCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUG .((((........))))((((((((...(((((((..((.((((((((((.(....))))))))))).)))))).)))...((((((...))))))......)))).)))) ( -31.70) >DroEre_CAF1 32559 111 - 1 ACGAGCUUUAAUCUUUGCGCAGCUGUUACACCGAGAAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUG (((((((.............((.((((((((((....((.((((((((((.(....))))))))))).))..)).)))....))))).)).............))))))). ( -25.67) >DroYak_CAF1 63231 111 - 1 ACGAGCUUUAAUCUUUGCGCAGCUGUUACAACGGAAAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUUGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCGGCUUGUG (((((((.........((((.(......)..(((...((.((((((((((.(....))))))))))).)).))).))))..((((((...)))))).......))))))). ( -27.70) >consensus ACGAGCUUUAAUCCUUGCGCAGCUGUUACACCGGCCAGAUAUUGGGUUAAUGCUGGCUUAACCCAAUUUCGCCGAGUGCCUUUGAUAACUUAUUAAUUAAAUCAGCUUGUG (((((((.........((((.((((......))))..((.((((((((((.(....))))))))))).)).....))))..((((((...)))))).......))))))). (-26.86 = -27.34 + 0.48)

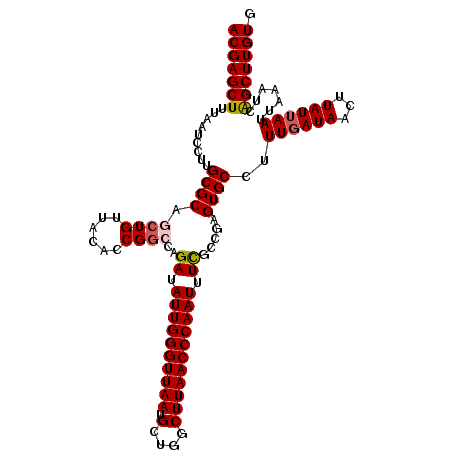
| Location | 19,398,444 – 19,398,548 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -16.76 |
| Energy contribution | -20.85 |
| Covariance contribution | 4.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19398444 104 + 23771897 CGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCACGGAUUAAAGCUCGU-------UAAGAAGGUGACUUCCGAGUUUCC------UAUCAAAUC .((.((((((((((.......)))))))))).)).((((((((((.....))))).)))....(((((((.-------...((((....)))))))))))))------......... ( -35.00) >DroSec_CAF1 37657 104 + 1 UGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCAAGGAUUAAAGCUCGU-------UAAGAAGGUGACUUCCGAGUUUCC------UAUCAAAUC .((.((((((((((.......)))))))))).))..((((((......)))).)).((((....((((((.-------...((((....)))))))))))))------)........ ( -34.40) >DroSim_CAF1 37546 104 + 1 CGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCAAGGAUUAAAGCUCGU-------UAAGAAGGUGACUUCCGAGUUUCC------UAUCAAAUC .((.((((((((((.......)))))))))).))..((((((......)))).)).((((....((((((.-------...((((....)))))))))))))------)........ ( -34.70) >DroEre_CAF1 32599 104 + 1 CGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUCUCGGUGUAACAGCUGCGCAAAGAUUAAAGCUCGU-------UAAGAAGGUGACUUCCGAGUUUCC------UAUCAAAUC .((.((((((((((.......)))))))))).)).(((..(((((.....)))))..)))...(((((((.-------...((((....)))))))))))..------......... ( -32.30) >DroYak_CAF1 63271 104 + 1 CAAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUUCCGUUGUAACAGCUGCGCAAAGAUUAAAGCUCGU-------UAAGAAGGUGACUUCCGAGUUUCC------UAUCAAAUC ....((((((((((.......))))))))))((((((.(((.((....)).))).))))))..(((((((.-------...((((....)))))))))))..------......... ( -31.40) >DroAna_CAF1 35492 112 + 1 --AAAGUGGGUUAAUCUAGCAUUAACCCCAUUUCCCCACAAUGCAA---CUGCGAAAGUAUAAAAGUUUAUGAAACCUUAAAGUGGAAAAUGCCAAUUCCCCAACUUAAAUCAAAUC --.((((((((((((.....))))))))(((((..((((.......---.(((....)))......((((........)))))))).)))))...........)))).......... ( -20.40) >consensus CGAAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCUGCGCAAGGAUUAAAGCUCGU_______UAAGAAGGUGACUUCCGAGUUUCC______UAUCAAAUC ....((((((((((.......)))))))))).........(((((.....)))))........(((((((...........((((....)))))))))))................. (-16.76 = -20.85 + 4.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:00 2006