
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,200,428 – 2,200,525 |
| Length | 97 |
| Max. P | 0.595131 |

| Location | 2,200,428 – 2,200,525 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
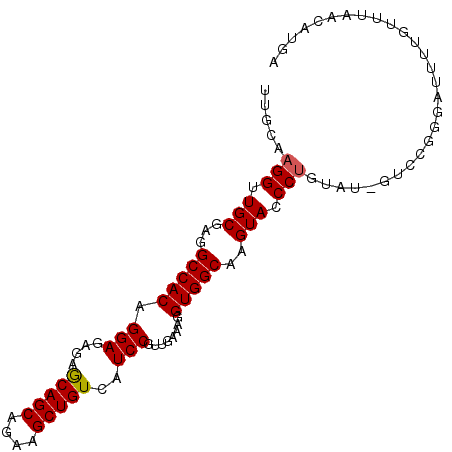
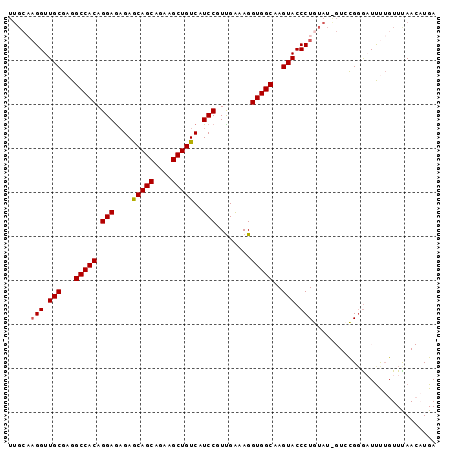
>3L_DroMel_CAF1 2200428 97 + 23771897 UUGCAAGGUUGCGAGGCCACAGGAGAGAGCAGCAGAAGCUGUCAUCCGUUGAAAGGUGGCAAGUACCCUGUAUGGUCCGCGAUUUUGUUUAACAUGA ..(((((((((((.(((((((((.((..(((((....)))))..)).(((.......)))......)))))..)))))))))))))))......... ( -34.40) >DroSec_CAF1 252424 97 + 1 UUGCAAGGUUGCGAGGCCACAGGAGAGAGCAGCAGAAGCUGUCAUCCGUUGAAAGGUGGCAAGUACCCUGUAUAGUCUGGGAUUUUGUUCAACAUGA ..((((((((((...(((((.(((....(((((....)))))..)))........)))))..)))(((.(......).))))))))))......... ( -26.90) >DroSim_CAF1 252935 97 + 1 UUGCAAGGUUGCGAGGCCACAGGAGAGAGCAGCAGAAGCUGUCAUCCGUUGAAAGGUGGCAAGUACCCUGUAUAGUCCGGGAUUUUGUUUAACAUGA ..((((((((.((.(((.(((((.((..(((((....)))))..)).(((.......)))......)))))...))))).))))))))......... ( -27.20) >DroEre_CAF1 257778 90 + 1 UUGG-AGGUUGCGAGGCCACAGGAAUGAACAGCAGAAGCUGUCAUCCGUUGGCAGGUGGCAAGUACCCUGU----UGCGGGUUUCGGUUUAA--UGA ((((-((.((((((.((((..(((.(((.((((....))))))))))..)))).((((.....))))...)----))))).)))))).....--... ( -30.10) >DroYak_CAF1 251472 85 + 1 ------GGUUGCGAGGCCACAGGAGAGAACAGCAGAAGCUGUCAUCCGUCGGCAGGUGGCAAGUACCCUGC----UGCGGAUUCCUGUUUAA--UGA ------((((....))))(((((((.((.((((....)))))).((((.(((((((.(.......))))))----)))))))))))))....--... ( -36.50) >consensus UUGCAAGGUUGCGAGGCCACAGGAGAGAGCAGCAGAAGCUGUCAUCCGUUGAAAGGUGGCAAGUACCCUGUAU_GUCCGGGAUUUUGUUUAACAUGA .....(((.(((...(((((.(((....(((((....)))))..)))........)))))..))).)))............................ (-23.42 = -23.38 + -0.04)

| Location | 2,200,428 – 2,200,525 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -17.08 |
| Energy contribution | -18.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
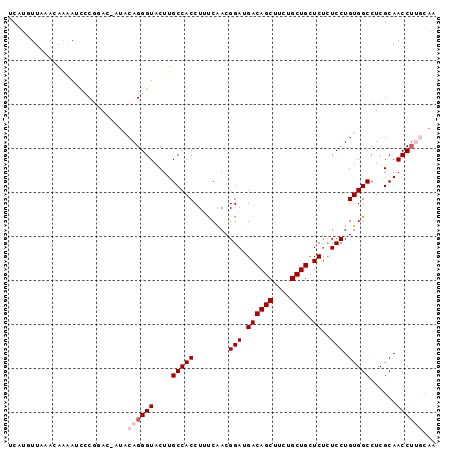
>3L_DroMel_CAF1 2200428 97 - 23771897 UCAUGUUAAACAAAAUCGCGGACCAUACAGGGUACUUGCCACCUUUCAACGGAUGACAGCUUCUGCUGCUCUCUCCUGUGGCCUCGCAACCUUGCAA .................(.((.(((((.(((((.......))))).....(((.((((((....)))).))..)))))))))).)(((....))).. ( -22.40) >DroSec_CAF1 252424 97 - 1 UCAUGUUGAACAAAAUCCCAGACUAUACAGGGUACUUGCCACCUUUCAACGGAUGACAGCUUCUGCUGCUCUCUCCUGUGGCCUCGCAACCUUGCAA ...........................((((((....(((((........(((.((((((....)))).))..))).)))))......))))))... ( -20.60) >DroSim_CAF1 252935 97 - 1 UCAUGUUAAACAAAAUCCCGGACUAUACAGGGUACUUGCCACCUUUCAACGGAUGACAGCUUCUGCUGCUCUCUCCUGUGGCCUCGCAACCUUGCAA ...........................((((((....(((((........(((.((((((....)))).))..))).)))))......))))))... ( -20.60) >DroEre_CAF1 257778 90 - 1 UCA--UUAAACCGAAACCCGCA----ACAGGGUACUUGCCACCUGCCAACGGAUGACAGCUUCUGCUGUUCAUUCCUGUGGCCUCGCAACCU-CCAA ...--..........((((...----...))))..((((.....((((..((((((((((....)))).))))))...))))...))))...-.... ( -27.00) >DroYak_CAF1 251472 85 - 1 UCA--UUAAACAGGAAUCCGCA----GCAGGGUACUUGCCACCUGCCGACGGAUGACAGCUUCUGCUGUUCUCUCCUGUGGCCUCGCAACC------ ...--....((((((((((((.----(((((..........))))).).)))))((((((....))))))...))))))............------ ( -31.40) >consensus UCAUGUUAAACAAAAUCCCGGAC_AUACAGGGUACUUGCCACCUUUCAACGGAUGACAGCUUCUGCUGCUCUCUCCUGUGGCCUCGCAACCUUGCAA ...........................((((((....(((((........(((.((((((....)))).))..))).)))))......))))))... (-17.08 = -18.08 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:32 2006