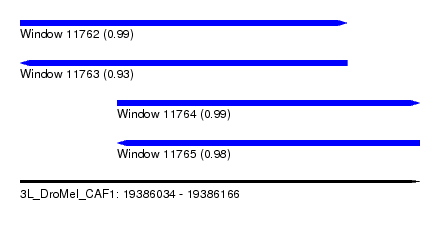
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,386,034 – 19,386,166 |
| Length | 132 |
| Max. P | 0.987511 |

| Location | 19,386,034 – 19,386,142 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -37.08 |
| Energy contribution | -36.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
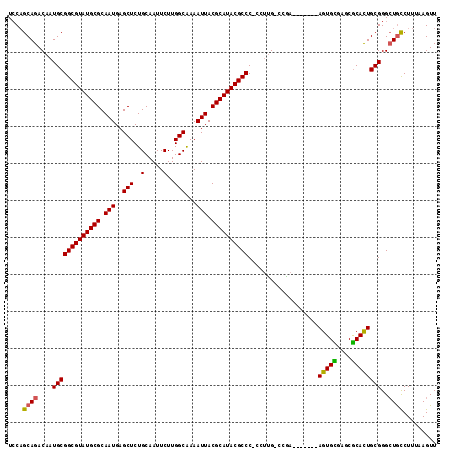
>3L_DroMel_CAF1 19386034 108 + 23771897 AACUUAAAGGCAGCCCGCAGUGCGCUCGCACU-------UCGG-CGAAGGGGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGA ..........((((..(((((((..((((...-------...)-)))....(((((((((((((((((((........)))))....)))))))))))))))))))))..)))).. ( -47.90) >DroSec_CAF1 18847 107 + 1 AACUUAAAGGCAGCCCGCAGUGCGCUCGCACU-------UCGG-CAAGG-GGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCAGGA ..(((..((((((((.(.(((((....)))))-------.)))-)...(-.(((((((((((((((((((........)).)))...)))))))))))))).)..)))))..))). ( -45.50) >DroSim_CAF1 21269 108 + 1 AACUUAAAGGCAGCCCGCAGUGCGCUCGCACU-------UCGG-CAAGGGGGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGA ........(((((.(((.(((((....)))))-------.)))-(((..(.(((((((((((((((((((........)).)))...)))))))))))))).).))).)))))... ( -45.00) >DroEre_CAF1 22867 107 + 1 AACUUUAAGGCAGCCCGCAGUGCGCUCACACU-------UUCG-CAAGG-GGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGA ..........((((..(((((((.........-------(((.-...))-)(((((((((((((((((((........)))))....)))))))))))))))))))))..)))).. ( -45.40) >DroYak_CAF1 53228 107 + 1 AACUUUAAGGCAGCCCGCAAUGCGCUCACACU-------UUCG-CAAGG-GGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGA ..........((((..(((((((.........-------(((.-...))-)(((((((((((((((((((........)))))....)))))))))))))))))))))..)))).. ( -45.40) >DroAna_CAF1 26099 114 + 1 AACUUAAAGAUA-CCCGCAAUGUGCACACACUCGUUCACUCGUACAUGG-GGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGUA .((....(((((-(((...((((((................))))))))-)(((((((((((((((((((........)).)))...))))))))))))))....)))))...)). ( -36.49) >consensus AACUUAAAGGCAGCCCGCAGUGCGCUCACACU_______UCGG_CAAGG_GGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGA ..........((((..((((((((....)......................((((((((((((((((......)))))))...((.....))))))))))))))))))..)))).. (-37.08 = -36.92 + -0.16)

| Location | 19,386,034 – 19,386,142 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -34.15 |
| Energy contribution | -33.73 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19386034 108 - 23771897 UCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCCCCUUCG-CCGA-------AGUGCGAGCGCACUGCGGGCUGCCUUUAAGUU ..((((.........((((((((((.(((..(((..(.....)..)))...))).)))))))))).......-(((.-------(((((....))))).))))))).......... ( -38.90) >DroSec_CAF1 18847 107 - 1 UCCUGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCC-CCUUG-CCGA-------AGUGCGAGCGCACUGCGGGCUGCCUUUAAGUU ....((((.(((.(.((((((((((.(((..(((..(.....)..)))...))).)))))))))).-).)))-(((.-------(((((....))))).))).))))......... ( -40.90) >DroSim_CAF1 21269 108 - 1 UCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCCCCCUUG-CCGA-------AGUGCGAGCGCACUGCGGGCUGCCUUUAAGUU ..((((.........((((((((((.(((..(((..(.....)..)))...))).)))))))))).......-(((.-------(((((....))))).))))))).......... ( -40.00) >DroEre_CAF1 22867 107 - 1 UCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCC-CCUUG-CGAA-------AGUGUGAGCGCACUGCGGGCUGCCUUAAAGUU ....((((.......((((((((((.(((..(((..(.....)..)))...))).)))))))))).-.((((-(...-------(((((....))))))))))))))......... ( -35.40) >DroYak_CAF1 53228 107 - 1 UCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCC-CCUUG-CGAA-------AGUGUGAGCGCAUUGCGGGCUGCCUUAAAGUU ..((((.(.((((((((((((((((.(((..(((..(.....)..)))...))).)))))))))).-.((..-((..-------..))..)).)))))))..)))).......... ( -38.50) >DroAna_CAF1 26099 114 - 1 UACAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCC-CCAUGUACGAGUGAACGAGUGUGUGCACAUUGCGGG-UAUCUUUAAGUU ....(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))(-(((..(((..(....)..)))..)((.....)))))-............ ( -35.70) >consensus UCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCC_CCUUG_CCGA_______AGUGCGAGCGCACUGCGGGCUGCCUUUAAGUU ....((((....(((((((((((((.(((..(((..(.....)..)))...))).))))))))))...................(((((....))))))))..))))......... (-34.15 = -33.73 + -0.42)

| Location | 19,386,066 – 19,386,166 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -30.76 |
| Energy contribution | -30.52 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
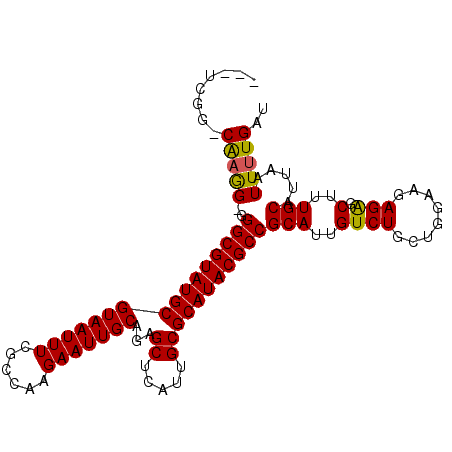
>3L_DroMel_CAF1 19386066 100 + 23771897 ---UCGG-CGAAGGGGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGAAGAGAGCUUUGCAUUAAUUUUGAU ---...(-((((((.(((((((((((((((((((........)))))....)))))))))))))).).((.(((......))).))))))))............ ( -38.00) >DroSec_CAF1 18879 99 + 1 ---UCGG-CAAGG-GGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCAGGAAGAGAGCUUUGCAUUAAUUUUGAU ---..((-(((.(-.(((((((((((((((((((........)).)))...)))))))))))))).).)))))((((((........))))))........... ( -36.00) >DroSim_CAF1 21301 100 + 1 ---UCGG-CAAGGGGGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGAAGAGAGCUUUGCAUUAAUUUUGAU ---...(-((((((.(((((((((((((((((((........)).)))...)))))))))))))).).((.(((......))).))))))))............ ( -36.00) >DroEre_CAF1 22899 99 + 1 ---UUCG-CAAGG-GGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGAAGAGAGCUUUGCAUUAAUUUUGAU ---...(-(((((-.(((((((((((((((((((........)))))....))))))))))))))...((.(((......))).))))))))............ ( -36.60) >DroYak_CAF1 53260 99 + 1 ---UUCG-CAAGG-GGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGAGGAGAGCUUUGCAUUAAUUUUGAU ---...(-(((((-.(((((((((((((((((((........)))))....))))))))))))))......(((.(....).))).))))))............ ( -36.90) >DroAna_CAF1 26134 103 + 1 CACUCGUACAUGG-GGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGUAAGAGGGCUUUGCAUUAAUUCUGAU ..((((....)))-)(((((((((((((((((((........)).)))...))))))))))))))(((..((((.(.....).))))..)))............ ( -34.80) >consensus ___UCGG_CAAGG_GGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGGAAGAGAGCUUUGCAUUAAUUUUGAU ........(((((..((((((((((((((((......)))))))...((.....)))))))))))(((..((((........))).)..))).....))))).. (-30.76 = -30.52 + -0.25)
| Location | 19,386,066 – 19,386,166 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
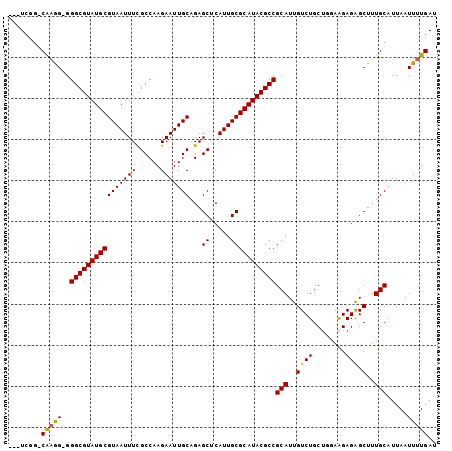
>3L_DroMel_CAF1 19386066 100 - 23771897 AUCAAAAUUAAUGCAAAGCUCUCUUCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCCCCUUCG-CCGA--- ............((.(((..........(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))..))).)-)...--- ( -26.30) >DroSec_CAF1 18879 99 - 1 AUCAAAAUUAAUGCAAAGCUCUCUUCCUGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCC-CCUUG-CCGA--- ............((((............(((.....)))((((((((((.(((..(((..(.....)..)))...))).)))))))))).-..)))-)...--- ( -28.00) >DroSim_CAF1 21301 100 - 1 AUCAAAAUUAAUGCAAAGCUCUCUUCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCCCCCUUG-CCGA--- ............((((............(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))....)))-)...--- ( -28.20) >DroEre_CAF1 22899 99 - 1 AUCAAAAUUAAUGCAAAGCUCUCUUCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCC-CCUUG-CGAA--- ...........(((((............(((.....)))((((((((((.(((..(((..(.....)..)))...))).)))))))))).-..)))-))..--- ( -28.30) >DroYak_CAF1 53260 99 - 1 AUCAAAAUUAAUGCAAAGCUCUCCUCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCC-CCUUG-CGAA--- ...........(((((............(((.....)))((((((((((.(((..(((..(.....)..)))...))).)))))))))).-..)))-))..--- ( -28.30) >DroAna_CAF1 26134 103 - 1 AUCAGAAUUAAUGCAAAGCCCUCUUACAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCC-CCAUGUACGAGUG ....................(((.(((((((.....)))((((((((((.(((..(((..(.....)..)))...))).)))))))))).-...)))).))).. ( -31.10) >consensus AUCAAAAUUAAUGCAAAGCUCUCUUCCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCC_CCUUG_CCGA___ ............................(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))............... (-24.62 = -24.62 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:48 2006