
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,384,789 – 19,384,895 |
| Length | 106 |
| Max. P | 0.744607 |

| Location | 19,384,789 – 19,384,895 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.74 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.63 |
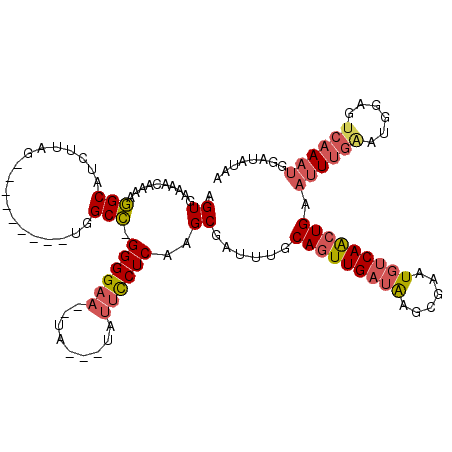
| Structure conservation index | 0.46 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
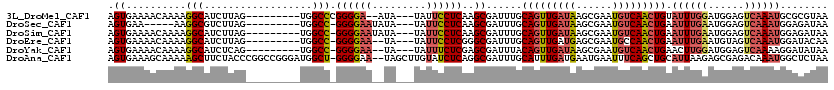
>3L_DroMel_CAF1 19384789 106 + 23771897 AGUGAAAACAAAAGGCAUCUUAG---------UGGCCCGGGGA--AUA---UAUUCCUCAAGCGAUUUGCAGUUGAUAAGCGAAUGUCAACUGUAUUUGGAUGGAGUCAAAUGCGCGUAA .((....)).....((((.....---------(((((((((((--(..---..))))))...(.(..(((((((((((......)))))))))))..).)..)).)))).))))...... ( -32.60) >DroSec_CAF1 17584 102 + 1 AGUGAA-----AAGGCGUCUUAG---------UGGCC-GGGGAAUAUA---UAUUCCUCAAGCGAUUUGCAGUUGAUAAGCGAAUGUCAACUGAAUUUGAAUGGAGUCAAAUGGAGAUAA ......-----.....(((((..---------(((((-(((((((...---.)))))))...(((....(((((((((......)))))))))...)))....).))))....))))).. ( -27.90) >DroSim_CAF1 20003 107 + 1 AGUGAAAACAAAAGGCAUCUUAG---------UGGCC-GGGGAAUAUA---UAUUCCUCAAGCGAUUUGCAGUUGAUAAGCGAAUGUCAACUGAAUUUGAAUGGAGUCAAAUGGAGAUAA .((....)).......(((((..---------(((((-(((((((...---.)))))))...(((....(((((((((......)))))))))...)))....).))))....))))).. ( -29.00) >DroEre_CAF1 21622 105 + 1 AGUGAAAACAAAAGGCAUCUUAG---------UGGCC-GGGGAA--UA---UAUUCCUCGGGCGAUUUGCAGUUGAUGAGCGAAUGCCAACUGAAUUUGAAUGUAGUCAAAUGGAUACAA .((....))....(((.......---------((.((-((((((--..---..)))))))).))((((((.........)))))))))......((((((......))))))........ ( -28.30) >DroYak_CAF1 51970 105 + 1 AGUGAAAACAAAAGGCAUCUCAG---------UGGCC-GGGGAA--UA---UAUUUCUCGAGCGAUUUACAGUUGAUAAGCGAAUGUCAACUGAACUUGGAUGGAGUCAAAAGGAUAUAA .((....))......(((((.((---------(.(((-((((((--..---..))))))).))......(((((((((......))))))))).))).)))))..(((.....))).... ( -28.90) >DroAna_CAF1 24836 117 + 1 AGUGAAAGCAAAAAGCUUCUACCCGGCCGGGAUGGCU-GGGGAA--UAGCUUGUAUCUCAGGCGAUUUGCAUUUGAUGAAUGAAUUUCAGCUGCAUUAAGAGCGAGACAAAUGGCUCUAA ((((..(((.....((.....(((((((.....))))-)))(((--(.(((((.....))))).))))))......((((.....))))))).)))).(((((.(......).))))).. ( -36.20) >consensus AGUGAAAACAAAAGGCAUCUUAG_________UGGCC_GGGGAA__UA___UAUUCCUCAAGCGAUUUGCAGUUGAUAAGCGAAUGUCAACUGAAUUUGAAUGGAGUCAAAUGGAUAUAA .((..........(((..................))).((((((.........))))))..))......(((((((((......))))))))).((((((......))))))........ (-14.10 = -14.74 + 0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:45 2006