
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,383,514 – 19,383,609 |
| Length | 95 |
| Max. P | 0.649077 |

| Location | 19,383,514 – 19,383,609 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -14.05 |
| Consensus MFE | -8.33 |
| Energy contribution | -8.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19383514 95 - 23771897 UAAACUAUAUCUUAUAGAU-UCAUUUGAAACAUG-----------------UAUA-UUUCUCCGUGGCCUCUUCUCUACUUUUAACUCCCUGCCACGGCACUUAGCCAACGCCA ....(((((...))))).(-((....))).....-----------------....-.......(((((.......................)))))(((.....)))....... ( -12.30) >DroSec_CAF1 16445 90 - 1 -----UAUAUCUUAUAGAU-UCAUUUGAAACAUG-----------------UAUA-UUUCUCAGUGGCCUCUUCUCUACUUUCAACUCCCUGCCACGGCACUUAGCCAACUCCA -----.......((((..(-((....)))...))-----------------))..-.......(((((.......................)))))(((.....)))....... ( -11.80) >DroSim_CAF1 18741 90 - 1 -----UAUAUCUUAUAGAU-UCAUUUGAAACAUG-----------------UUUA-UUUCUCCGUGGCCUCUUCUCUACUUUCAACUCCCUGCGACGGCACUUAGCCAACGCCA -----.........(((((-((....))).((((-----------------....-......))))........)))).............(((..(((.....)))..))).. ( -11.60) >DroEre_CAF1 20328 95 - 1 UCUUUUAUAUUUGAAAUAU-UCAUUUUACACAUG-----------------AAUA-UUUCUCCGUGGCCUCUUCUCUACUUUCAACUCCCUGCCACGGCACUUAGCCAACGCCA ............(((((((-((((.......)))-----------------))))-))))...(((((.......................)))))(((.....)))....... ( -20.60) >DroYak_CAF1 50723 95 - 1 UUUU-UUUAUCUGAAAUAU-UAAUUUUAAACGUG-----------------AAUUUUUUCUCCGUGGCCUCUUCUCCACUUUUAACUCCCUGCCACGGCACUUAGCCAACGCCA ....-......((((((..-..))))))...(((-----------------............(((((.......................)))))(((.....)))..))).. ( -13.50) >DroAna_CAF1 23053 106 - 1 -----GAUGUUUAAUAGAUAGCAGUUGAAACUUUAAUAAACUAUAUAAGUUGAUUUUCUCUACGUG---UAUGCUUUGGUCCUAACUCCCUGCCACGGCACUUAGCCAACGCCA -----(.((((.........((((..((........((((.((((((.((.((.....)).)).))---)))).))))........)).))))...(((.....))))))).). ( -14.49) >consensus _____UAUAUCUUAUAGAU_UCAUUUGAAACAUG_________________UAUA_UUUCUCCGUGGCCUCUUCUCUACUUUCAACUCCCUGCCACGGCACUUAGCCAACGCCA ...............................................................(((((.......................)))))(((.....)))....... ( -8.33 = -8.83 + 0.50)

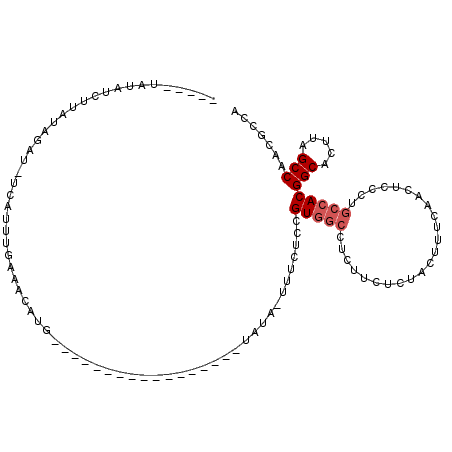

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:43 2006