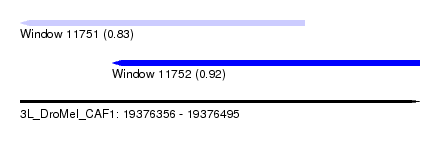
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,376,356 – 19,376,495 |
| Length | 139 |
| Max. P | 0.916701 |

| Location | 19,376,356 – 19,376,455 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -14.11 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19376356 99 - 23771897 GGCACU--UUGUUUGC-----UCU-ACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCU--UGUGUUGUC--------UUCUGGCGUGUUCUGCCGUAUUUUACUCUGCU--- ((((.(--((((((((-----...-..)))....)))))).....(((((..((((((((((....--)))))))))--------)..))).))....))))...............--- ( -27.50) >DroSec_CAF1 9257 99 - 1 GGCACU--UUGUUUGC-----UCU-ACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCU--UGUGUUGUC--------UUCUGGCUUGUUCUGCCGUAUUUUACUCUGCU--- ((((.(--((((((((-----...-..)))....)))))).....(((....((((((((((....--)))))))))--------)...)))......))))...............--- ( -26.70) >DroSim_CAF1 6635 102 - 1 GGCACU--UUGUUUGC-----UCU-ACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCU--UGUGUUGUC--------UUCUGGCUUGUUCUGCCGUAUUUUACUCUGCUCUG ((((.(--((((((((-----...-..)))....)))))).....(((....((((((((((....--)))))))))--------)...)))......)))).................. ( -26.70) >DroEre_CAF1 13229 99 - 1 GGCACU--UUGUUUGC-----UCU-ACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCU--UGUGUUGUC--------UUCUGGGCUGUUCUGCCGUAUUUUACUCUGCU--- ((((.(--((((((((-----...-..)))....)))))).....(((((..((((((((((....--)))))))))--------)..))))).....))))...............--- ( -33.00) >DroWil_CAF1 30771 117 - 1 UGUAGUUGUUGUUUGCCUACUUCUAACGCAUUACGGCAAAUAUUUGCGCAAUGGACAACAUAUUCUCUUAGGUUGUAUGUCUGUGUACUUACUUAUCUUUCCGUUUUCUAUUCUGUU--- .((((....((((((((((...(....)...)).)))))))).))))(.((((((.(((.........(((((.((((......))))..))))).......))).)))))))....--- ( -19.59) >DroAna_CAF1 15792 98 - 1 GGCACU--CUGUUUGC-----UUU-ACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCU--UGUGUUGUC--------UUCUGGGUUUUUUU-UUUUGGUUUAUUCGGUC--- (((...--.(((.(((-----...-..)))..))).((((.....(((((..((((((((((....--)))))))))--------)..)))))......-.)))).........)))--- ( -26.30) >consensus GGCACU__UUGUUUGC_____UCU_ACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCU__UGUGUUGUC________UUCUGGCUUGUUCUGCCGUAUUUUACUCUGCU___ ((((....((((((((...........)))....))))).....)))).....(((((((((......)))))))))........................................... (-14.11 = -13.92 + -0.19)

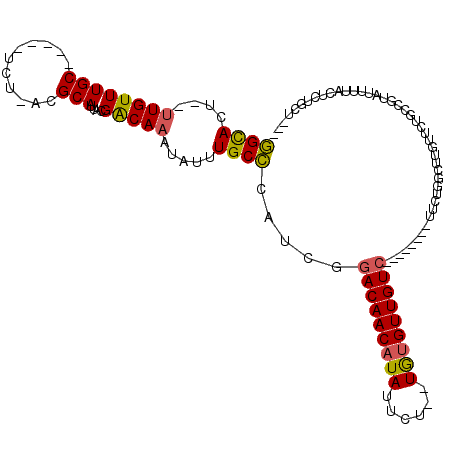
| Location | 19,376,388 – 19,376,495 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19376388 107 - 23771897 CCGCUUCGAGACUUUCCCCAAAUAUAUUUCCAUCAUCAGAGGCACUUUGUUUGCUCUACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCUUGUGUUGUC ......(((.......................((....))((((.(((((((((.....)))....))))))....))))..)))(((((((((....))))))))) ( -20.10) >DroSec_CAF1 9289 107 - 1 CCGUUUCGAGUCUUUCCCCAAAUAUAUUUCCAUCAUCAGAGGCACUUUGUUUGCUCUACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCUUGUGUUGUC ......(((.......................((....))((((.(((((((((.....)))....))))))....))))..)))(((((((((....))))))))) ( -20.10) >DroSim_CAF1 6670 107 - 1 CCGCUUCGAGUCUUUCCCCAAAUAUAUUUCCAUCAUCAGAGGCACUUUGUUUGCUCUACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCUUGUGUUGUC ......(((.......................((....))((((.(((((((((.....)))....))))))....))))..)))(((((((((....))))))))) ( -20.10) >DroEre_CAF1 13261 107 - 1 CCGCUUCGAGACUUUCCCCAAAUAUAUUUCCACCAUCAGAGGCACUUUGUUUGCUCUACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCUUGUGUUGUC ......(((...................((........))((((.(((((((((.....)))....))))))....))))..)))(((((((((....))))))))) ( -19.50) >DroYak_CAF1 43598 107 - 1 CCGCUUCGAGACUUUCCCCAAAUAUAUUUCCAUCAUCAGAGGCACUUUGUUUGCUCUACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCUUGUGUUGUC ......(((.......................((....))((((.(((((((((.....)))....))))))....))))..)))(((((((((....))))))))) ( -20.10) >DroAna_CAF1 15823 107 - 1 GCUCUCGGAGACUUUUCCCAAAUAUAUUCCCAUCAUCUCCGGCACUCUGUUUGCUUUACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCUUGUGUUGUC ......(((((........................)))))((((...(((((((.....)))....))))......)))).....(((((((((....))))))))) ( -21.56) >consensus CCGCUUCGAGACUUUCCCCAAAUAUAUUUCCAUCAUCAGAGGCACUUUGUUUGCUCUACGCAUUACGACAAAUAUUUGCCCAUCGGACAACAUAUUCUUGUGUUGUC ........................................((((.(((((((((.....)))....))))))....)))).....(((((((((....))))))))) (-18.55 = -18.72 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:36 2006