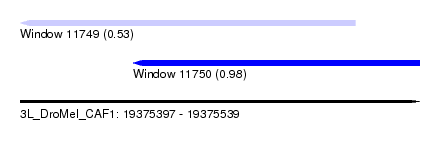
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,375,397 – 19,375,539 |
| Length | 142 |
| Max. P | 0.983901 |

| Location | 19,375,397 – 19,375,516 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -23.65 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
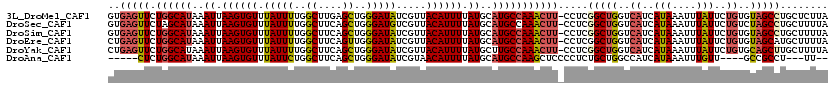
>3L_DroMel_CAF1 19375397 119 - 23771897 GUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUGAGCUGGGAUAUCGUUACAUUUUAUGCAUGCCAAACUU-CCUCGGCUGGUCAUCAUAAAUUUAUUCUGUGUAGCCUGCUCUUA (..((((.((((((..((.((((((.(((((..((....))..))))).....)))))).))..)))))))))).-.)..(((.(((...((((.........))))..))).))).... ( -31.50) >DroSec_CAF1 8326 119 - 1 GUGAGUUCUAGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUGUCGUUACAUUUUAUGCAUGCCAAACUU-CCUCGGCUGGUCAUCAUAAAUUUAUUCUGUCUAGCCUGCUUUUA (..((((...((((..((.((((((.(((((..((....))..))))).....)))))).))..))))..)))).-.)..((((((.((..(((....)))..)).))))))........ ( -29.40) >DroSim_CAF1 5724 119 - 1 GUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUAUCGUUACAUUUUAUGCAUGCCAAACUU-CCUCGGCUGGUCAUCAUAAAUUUAUUCUGUGUAGCCUGCUUUUA (..((((.((((((..((.((((((.(((((..((....))..))))).....)))))).))..)))))))))).-.)..(((.(((...((((.........))))..))).))).... ( -31.50) >DroEre_CAF1 12200 119 - 1 CUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGUUGGGAUAUCGUUACAUUUUAUGCAUGCCAAACUU-CCUCGGCUGGUCAUCAUAAAUUUAUUCUGUGUAGCAUGCUUUUA .........(((((............(((((..((....))..)))))..(((((((((((((.((((((..(..-....)..))).)))))))))........)))))))))))).... ( -25.90) >DroYak_CAF1 41301 119 - 1 CUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUAUCGUUACAUUUUAUGCUUGCCAAACUU-CCUCGGCUGGUCAUCAUAAAUUUAUUCUGUGCAGCUUGCUUUUA ..(((((.(((((...((.((((((.(((((..((....))..))))).....)))))).))...))))))))))-....(((((..((..(((....)))..))..)))))........ ( -27.60) >DroAna_CAF1 15215 106 - 1 -----CUCUGGCAUAAAUUAAGUGUUUAUUCUGGCUUCAGCUGGGAUAUCGUAACAUUUUAUGCAUGCCAAGCUCCCCUCUGCUGGCCAUCAUAAAUUUGUU----GCCGCCU---UU-- -----....(((..............(((((..((....))..)))))..((((((.((((((.((((((.((........))))).)))))))))..))))----)).))).---..-- ( -32.80) >consensus GUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUAUCGUUACAUUUUAUGCAUGCCAAACUU_CCUCGGCUGGUCAUCAUAAAUUUAUUCUGUGUAGCCUGCUUUUA ..(((((.((((((..((.((((((.(((((..((....))..))))).....)))))).))..))))))))))).....(((((..((..(((....)))..))..)))))........ (-23.65 = -24.18 + 0.53)


| Location | 19,375,437 – 19,375,539 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.96 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19375437 102 - 23771897 -----------------ACAUUCACACUCAUGGACUAGUUGUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUGAGCUGGGAUAUCGUUACAUUUUAUGCAUGCCAAACUU-CCUC -----------------........(((((..(.....)..)))))..((((((..((.((((((.(((((..((....))..))))).....)))))).))..)))))).....-.... ( -26.30) >DroSec_CAF1 8366 102 - 1 -----------------ACACUCGCACUCAUGGACUGGUUGUGAGUUCUAGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUGUCGUUACAUUUUAUGCAUGCCAAACUU-CCUC -----------------..(((((((..((.....))..)))))))....(((((((....(((..(((((..((....))..)))))....)))..)))))))...........-.... ( -25.00) >DroSim_CAF1 5764 102 - 1 -----------------ACACUCACACUCACGGACUGGUUGUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUAUCGUUACAUUUUAUGCAUGCCAAACUU-CCUC -----------------..(((((((..((.....))..)))))))..((((((..((.((((((.(((((..((....))..))))).....)))))).))..)))))).....-.... ( -30.00) >DroYak_CAF1 41341 119 - 1 CCAACACCCACACACUCGCACUCACACUCAUGGACUAUUUCUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUAUCGUUACAUUUUAUGCUUGCCAAACUU-CCUC .........................(((((.(((....))))))))..(((((...((.((((((.(((((..((....))..))))).....)))))).))...))))).....-.... ( -23.80) >DroAna_CAF1 15246 80 - 1 ------------------------CACUC----------------CUCUGGCAUAAAUUAAGUGUUUAUUCUGGCUUCAGCUGGGAUAUCGUAACAUUUUAUGCAUGCCAAGCUCCCCUC ------------------------.....----------------...((((((..((.((((((((((((..((....))..)))))....))))))).))..)))))).......... ( -22.40) >consensus _________________ACACUCACACUCAUGGACUAGUUGUGAGUUCUGGCAUAAAUUAAGUGUUUAUUUUGGCUUCAGCUGGGAUAUCGUUACAUUUUAUGCAUGCCAAACUU_CCUC .........................((((((((.....))))))))..((((((..((.((((((.(((((..((....))..))))).....)))))).))..)))))).......... (-20.76 = -21.96 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:34 2006