
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,362,893 – 19,363,009 |
| Length | 116 |
| Max. P | 0.894380 |

| Location | 19,362,893 – 19,363,009 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -27.82 |
| Energy contribution | -27.71 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
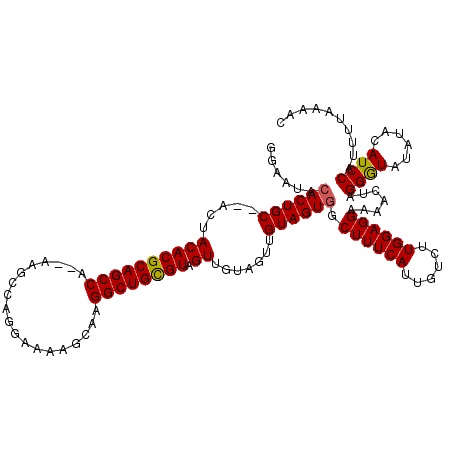
>3L_DroMel_CAF1 19362893 116 + 23771897 GGAAUACACUGC--ACUACACGCAGCCG--AAGCCAGGAAAAGCAAGGCUGCGUAGUUGUAGUUGUAGUUGCUUUCAUUGUCUUGGAGGAAAACUAGGGUGUAUACAUCCAUUUUAAAAC ((((((((((..--(((((((((((((.--..((........))..))))))))....)))))..(((((.((((((......))))))..))))).)))))))...))).......... ( -38.10) >DroSim_CAF1 13294 118 + 1 GGAAUACACUGCACACUACACGCAGCCC--GAGCCAGGAAAAGCGUGGCUGUGUAGUUGUAGUUGUAGUGGCUUUCAUUGUCUUGGAGGAAAACUAGGGUAUAUACAUCCAUUUUAAAAC (((...(((((((.(((((((((((((.--..((........))..))))))))....)))))))))))).((((((......))))))..................))).......... ( -37.20) >DroYak_CAF1 16927 109 + 1 GGAAUACACUGC--ACUACACGCAGCCAAGAAUACAGG---AGCAAGGCUGCGUAGUU------GUAGUGGCUUUCAUUGUCUUGGAGGAAAACUAGGAUAUAUACAUCCAUUUUAAAAC (((...((((((--(((..((((((((...........---.....)))))))))).)------))))))........((((((((.......))))))))......))).......... ( -33.39) >consensus GGAAUACACUGC__ACUACACGCAGCCA__AAGCCAGGAAAAGCAAGGCUGCGUAGUUGUAGUUGUAGUGGCUUUCAUUGUCUUGGAGGAAAACUAGGGUAUAUACAUCCAUUUUAAAAC ......((((((.....((((((((((...................)))))))).)).......)))))).((((((......)))))).......((((......)))).......... (-27.82 = -27.71 + -0.11)


| Location | 19,362,893 – 19,363,009 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
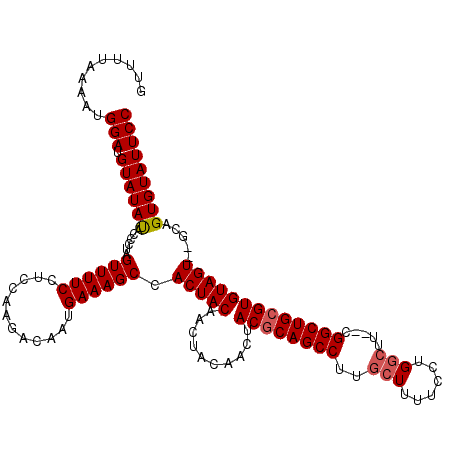
>3L_DroMel_CAF1 19362893 116 - 23771897 GUUUUAAAAUGGAUGUAUACACCCUAGUUUUCCUCCAAGACAAUGAAAGCAACUACAACUACAACUACGCAGCCUUGCUUUUCCUGGCUU--CGGCUGCGUGUAGU--GCAGUGUAUUCC ..........(((.((((((......((((((............)))))).......(((((....((((((((..(((......)))..--.)))))))))))))--...))))))))) ( -32.80) >DroSim_CAF1 13294 118 - 1 GUUUUAAAAUGGAUGUAUAUACCCUAGUUUUCCUCCAAGACAAUGAAAGCCACUACAACUACAACUACACAGCCACGCUUUUCCUGGCUC--GGGCUGCGUGUAGUGUGCAGUGUAUUCC ..........(((.((((((......(((((.....))))).......((((((((..........((.(((((..(((......)))..--.))))).)))))))).)).))))))))) ( -28.80) >DroYak_CAF1 16927 109 - 1 GUUUUAAAAUGGAUGUAUAUAUCCUAGUUUUCCUCCAAGACAAUGAAAGCCACUAC------AACUACGCAGCCUUGCU---CCUGUAUUCUUGGCUGCGUGUAGU--GCAGUGUAUUCC ..........(((.((((((......((((((............))))))((((((------....((((((((.(((.---...))).....)))))))))))))--)..))))))))) ( -29.30) >consensus GUUUUAAAAUGGAUGUAUAUACCCUAGUUUUCCUCCAAGACAAUGAAAGCCACUACAACUACAACUACGCAGCCUUGCUUUUCCUGGCUU__CGGCUGCGUGUAGU__GCAGUGUAUUCC ..........(((.((((((......((((((............)))))).(((((..........((((((((..(((......))).....))))))))))))).....))))))))) (-22.82 = -23.60 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:29 2006