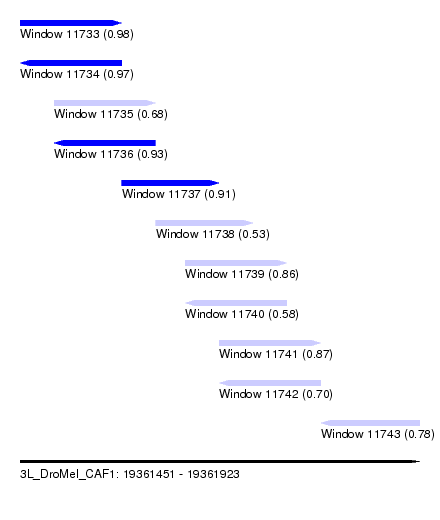
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,361,451 – 19,361,923 |
| Length | 472 |
| Max. P | 0.980625 |

| Location | 19,361,451 – 19,361,571 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -33.01 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361451 120 + 23771897 UGUUCGAGGGACUUAGUUUGUAUUGGAAACAAACGUUUGCUUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCCCAAGU .....(((((.....((((((.......)))))).....)))))..((((.((((.(((((((((.((....))....((((.............))))..))))))))))))).)))). ( -32.12) >DroSec_CAF1 11160 120 + 1 UGUUCGGGGGAUUUAGUUUGUGUUGGAAACAAACGUUCGCUUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCUCAAGU .....(((.((....((((((.......))))))..)).)))....((((.((((.(((((((((.((....))....((((.............))))..))))))))))))).)))). ( -33.82) >DroSim_CAF1 11454 120 + 1 UGUUCGGGGGAUUUAGUUUGUGUUGGAAACAAACGUUCGCUCUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCCCAAGU .....(((.((....((((((.......))))))..)).)))....((((.((((.(((((((((.((....))....((((.............))))..))))))))))))).)))). ( -35.62) >DroYak_CAF1 15480 120 + 1 UGUUUGGGGGAUUUAGUUUGUGCUGGAAACAAACGUUUGCUUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCUGCCCCAAGU ..(((((((......((((((.......))))))....(((..........)))..(((((((((.((....))....((((.............))))..)))))))))..))))))). ( -31.22) >consensus UGUUCGGGGGAUUUAGUUUGUGUUGGAAACAAACGUUCGCUUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCCCAAGU .....(((.((....((((((.......))))))..)).)))....((((.((((.(((((((((.((....))....((((.............))))..))))))))))))).)))). (-33.01 = -32.20 + -0.81)


| Location | 19,361,451 – 19,361,571 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -28.69 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361451 120 - 23771897 ACUUGGGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAAGCAAACGUUUGUUUCCAAUACAAACUAAGUCCCUCGAACA (((((((((((((((((((.((((((......((((.....))))...)))))).))))))))).)))).........((((((....))))))..........)))))).......... ( -31.40) >DroSec_CAF1 11160 120 - 1 ACUUGAGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAAGCGAACGUUUGUUUCCAACACAAACUAAAUCCCCCGAACA ..(((.(((((((((((((.((((((......((((.....))))...)))))).))))))))).)))).))).............((((((.......))))))............... ( -30.00) >DroSim_CAF1 11454 120 - 1 ACUUGGGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAGAGCGAACGUUUGUUUCCAACACAAACUAAAUCCCCCGAACA ..(((((((((((((((((.((((((......((((.....))))...)))))).))))))))).)))).................((((((.......))))))........))))... ( -32.10) >DroYak_CAF1 15480 120 - 1 ACUUGGGGCAGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAAGCAAACGUUUGUUUCCAGCACAAACUAAAUCCCCCAAACA ..((((((..(((((((((.((((((......((((.....))))...)))))).)))))))))......................((((((.......))))))......))))))... ( -29.70) >consensus ACUUGGGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAAGCAAACGUUUGUUUCCAACACAAACUAAAUCCCCCGAACA ..(((((((((((((((((.((((((......((((.....))))...)))))).))))))))).)))).................((((((.......))))))........))))... (-28.69 = -28.88 + 0.19)


| Location | 19,361,491 – 19,361,611 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -31.37 |
| Energy contribution | -31.25 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361491 120 + 23771897 UUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCCCAAGUAAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUU ((((((((.(.((((.(((((((((.((....))....((((.............))))..)))))))))))))).))))))))..........((((((.........))))))..... ( -32.62) >DroSec_CAF1 11200 120 + 1 UUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCUCAAGUAAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUU ((((((((.(.((((.(((((((((.((....))....((((.............))))..))))))))))))).)))))))))..........((((((.........))))))..... ( -32.82) >DroSim_CAF1 11494 120 + 1 UCUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCCCAAGUAAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUU ..((((((.(.((((.(((((((((.((....))....((((.............))))..)))))))))))))).))))))............((((((.........))))))..... ( -31.52) >DroYak_CAF1 15520 120 + 1 UUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCUGCCCCAAGUAAAGCUGGCUUAAAGACCCGGGCUCCGAAUGGGUUACGUU ((((((((.(.((((.(((((((((.((....))....((((.............))))..)))))))))))))).))))))))..........((((((..(...)..))))))..... ( -30.72) >consensus UUUUAUUUUGUGGCGUGCCAUUGCAACUAAAUGGAAAAUGAAAAAUUUAUAGAAAUUCAAAUGCAAUGGCCGCCCCAAGUAAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUU ((((((((.(.((((.(((((((((.((....))....((((.............))))..)))))))))))))).))))))))..........((((((.........))))))..... (-31.37 = -31.25 + -0.13)



| Location | 19,361,491 – 19,361,611 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -29.62 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361491 120 - 23771897 AACGUAACCCAUUCGGAACCCGGGUCUUUAAGCCAGCUUUACUUGGGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAA ........((....))...((((((....(((....))).))))))(((((((((((((.((((((......((((.....))))...)))))).))))))))).))))........... ( -32.20) >DroSec_CAF1 11200 120 - 1 AACGUAACCCAUUCGGAACCCGGGUCUUUAAGCCAGCUUUACUUGAGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAA ...((((..(...(((...)))(((......))).)..))))(((.(((((((((((((.((((((......((((.....))))...)))))).))))))))).)))).)))....... ( -31.30) >DroSim_CAF1 11494 120 - 1 AACGUAACCCAUUCGGAACCCGGGUCUUUAAGCCAGCUUUACUUGGGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAGA ........((....))...((((((....(((....))).))))))(((((((((((((.((((((......((((.....))))...)))))).))))))))).))))........... ( -32.20) >DroYak_CAF1 15520 120 - 1 AACGUAACCCAUUCGGAGCCCGGGUCUUUAAGCCAGCUUUACUUGGGGCAGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAA ..(((..((((...(((((...(((......))).)))))...))))...(((((((((.((((((......((((.....))))...)))))).))))))))))))............. ( -31.80) >consensus AACGUAACCCAUUCGGAACCCGGGUCUUUAAGCCAGCUUUACUUGGGGCGGCCAUUGCAUUUGAAUUUCUAUAAAUUUUUCAUUUUCCAUUUAGUUGCAAUGGCACGCCACAAAAUAAAA ...((((..(...(((...)))(((......))).)..))))(((.(((((((((((((.((((((......((((.....))))...)))))).))))))))).)))).)))....... (-29.62 = -29.88 + 0.25)


| Location | 19,361,571 – 19,361,686 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.05 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361571 115 + 23771897 AAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUUGCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGG-----ACUGGAAACUGGAGGGAAACCGAGAGCUAUCAAUGGCAGCA ...((((.(........((((.(((((..(.((((...((((....)))).(((..........))).)))).)-----..))))).)))).((....)).............).)))). ( -34.00) >DroSec_CAF1 11280 120 + 1 AAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUUGCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGGACUGGACUGGAAGCUGGUGGGAAACCGAGAGCUAUCAAUGGCAGCA ...((((.(........((((.(((((.(((..(((..((((....))))..)))....)))..))))).)))).......(((.((((..(((....)))..)))).)))..).)))). ( -41.10) >DroSim_CAF1 11574 120 + 1 AAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUUGCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGGACUGGACUGGAAACUGGUGGGAAACCGAGAGCUAUCAAUGGCAGCA ...((((.(........((((.(((((..(.((((...((((....))))...............(....)..)))).)..))))).))))(((....)))............).)))). ( -35.00) >DroYak_CAF1 15600 111 + 1 AAAGCUGGCUUAAAGACCCGGGCUCCGAAUGGGUUACGUUACGGAAUCAAACGGAAAAAUAUAGCGGAGACUGGACUG-----GAA----GAGGGAAACUGAGAGCUAUCAAUGGCAGCA ...((((.(........((((..((((((((.....)))).))))..)...)))......((((((....).......-----...----..((....))....)))))....).)))). ( -28.80) >consensus AAAGCUGGCUUAAAGACCCGGGUUCCGAAUGGGUUACGUUGCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGGACUG_ACUGGAAACUGGAGGGAAACCGAGAGCUAUCAAUGGCAGCA ...((((.(........((((.(((((.(((..((...((((....))))....))...)))..))))).))))..................((....)).............).)))). (-28.92 = -29.05 + 0.13)



| Location | 19,361,611 – 19,361,726 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361611 115 + 23771897 GCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGG-----ACUGGAAACUGGAGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUG ((....))...........(((((((((...(((-----..(((.(((((..((....))....((((....))))..))))).))).......)))..))))).))))........... ( -26.50) >DroSec_CAF1 11320 120 + 1 GCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGGACUGGACUGGAAGCUGGUGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUG ((....))...........(((((((((...(((.......(((.((((..(((....)))..)))).))).((((((...)))))).......)))..))))).))))........... ( -30.20) >DroSim_CAF1 11614 120 + 1 GCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGGACUGGACUGGAAACUGGUGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUG ((....))...........(((((((((...(((.......(((.(((((.(((....)))...((((....))))..))))).))).......)))..))))).))))........... ( -28.54) >DroYak_CAF1 15640 111 + 1 ACGGAAUCAAACGGAAAAAUAUAGCGGAGACUGGACUG-----GAA----GAGGGAAACUGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUG ............((.......(((((....)..(((((-----...----..((....))....((((....))))..))))))))).......))...(((((.......))))).... ( -20.24) >consensus GCGGAAGCAAACUGAAAAAUAUAGCGGAGACUGGACUG_ACUGGAAACUGGAGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUG ......(((((.........((((((....).....................((....))....)))))...(((((...((....))....)))))..)))))................ (-20.54 = -20.60 + 0.06)

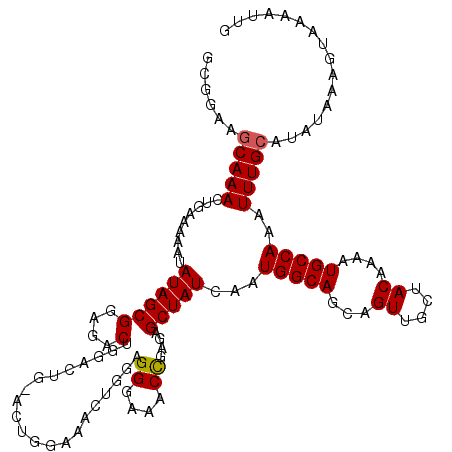

| Location | 19,361,646 – 19,361,766 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -21.71 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361646 120 + 23771897 CUGGAAACUGGAGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGUGGGUGUAGCCAGGGGGGAGGGGG (((..(((((..((....))....((((....))))..)))))((((((......(((.(((((.......))))).)))......((((.....)))).)))))))))........... ( -31.90) >DroSec_CAF1 11360 119 + 1 CUGGAAGCUGGUGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGGGUGGGGAAACCGGAGGGGG-AGG .(((.((((..(((....)))..)))).))).(((((.(((((..(((...((((.......)))).....))).)))))....)))))((.((....(.((....)).)...)).-)). ( -36.00) >DroSim_CAF1 11654 120 + 1 CUGGAAACUGGUGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGGGUGGGGAAGCCGGAGGGGGAAGG .......(((((((....)).....(((.((.(((((.(((((..(((...((((.......)))).....))).)))))....))))).)).))).........))))).......... ( -28.80) >DroYak_CAF1 15678 98 + 1 ---GAA----GAGGGAAACUGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGGUAGUGGGC---------------UGGG ---...----..((....)).((.(((((((.(((((.(((((..(((...((((.......)))).....))).)))))....))))).)))))))...)---------------)... ( -26.10) >consensus CUGGAAACUGGAGGGAAACCGAGAGCUAUCAAUGGCAGCAGUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGGGGGGGGAA_CCGGAGGGGG_AGG .......(((((((....)).....(((.((.(((((.(((((..(((...((((.......)))).....))).)))))....))))).)).))).........))))).......... (-21.71 = -22.65 + 0.94)

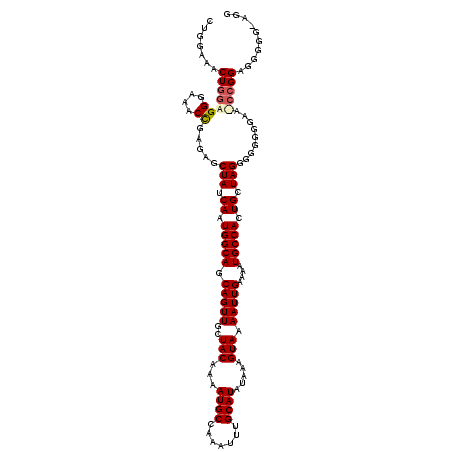
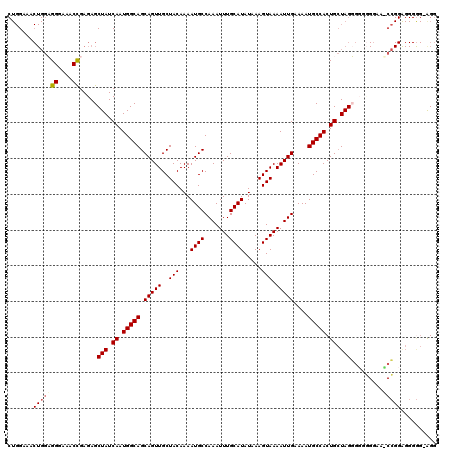
| Location | 19,361,646 – 19,361,766 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361646 120 - 23771897 CCCCCUCCCCCCUGGCUACACCCACUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAACUGCUGCCAUUGAUAGCUCUCGGUUUCCCUCCAGUUUCCAG ...........((((..((.....(((.((((((((...................((((((........))))))((....)))))))))).))).....((.......)).))..)))) ( -25.40) >DroSec_CAF1 11360 119 - 1 CCU-CCCCCUCCGGUUUCCCCACCCUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAACUGCUGCCAUUGAUAGCUCUCGGUUUCCCACCAGCUUCCAG ...-........((...((.....(((.((((((((...................((((((........))))))((....)))))))))).))).....))...))............. ( -22.00) >DroSim_CAF1 11654 120 - 1 CCUUCCCCCUCCGGCUUCCCCACCCUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAACUGCUGCCAUUGAUAGCUCUCGGUUUCCCACCAGUUUCCAG ............((...((.....(((.((((((((...................((((((........))))))((....)))))))))).))).....))....))............ ( -21.50) >DroYak_CAF1 15678 98 - 1 CCCA---------------GCCCACUACCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAACUGCUGCCAUUGAUAGCUCUCAGUUUCCCUC----UUC--- ....---------------.....(((.((((((((...................((((((........))))))((....)))))))))).)))...............----...--- ( -18.30) >consensus CCC_CCCCCUCCGG_UUCCCCACACUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAACUGCUGCCAUUGAUAGCUCUCGGUUUCCCACCAGUUUCCAG ........................(((.((((((((...................((((((........))))))((....)))))))))).)))......................... (-18.20 = -18.20 + -0.00)

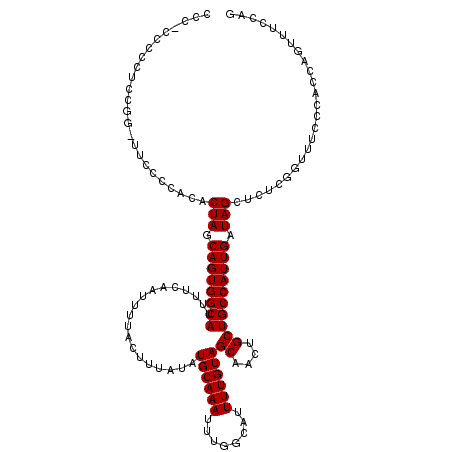

| Location | 19,361,686 – 19,361,806 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -19.11 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361686 120 + 23771897 GUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGUGGGUGUAGCCAGGGGGGAGGGGGAGGCAGCAGAAAGCAACCGAAAAAAGGAAAAUCGGAAGGG ((((((......((((...(((((.......)))))........(.((.((.((..(((......))).)).)).)).)..))))......))))))..............((....)). ( -26.60) >DroSec_CAF1 11400 119 + 1 GUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGGGUGGGGAAACCGGAGGGGG-AGGGGGCGGCAGAAAGCAACCGAAAAAAGGAAAAUCGGAAGGG ((((((......((((...(((((.......))))).........(((.((.((....(.((....)).)...)).-))..)))))))...))))))..............((....)). ( -32.10) >DroSim_CAF1 11694 120 + 1 GUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGGGUGGGGAAGCCGGAGGGGGAAGGGGGCGGCAGAAAGCAACCGAAAAAAGGAAAAUCGGAAGGG ((((((......((((...(((((.......))))).........(((.((.((..(((......)))..)).))......)))))))...))))))..............((....)). ( -28.30) >DroYak_CAF1 15711 105 + 1 GUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGGUAGUGGGC---------------UGGGGGGCAGCAGAAACCAACCGAAAAAAGGAAAAUCGGAAAGG (((((..(.....(((((.(((((.......))))).)))......((((.....))))))---------------...)..))))).........((((...........))))..... ( -22.20) >consensus GUUGCUACAAAAUGCCAAAUUUGCAUAUAAAGUAAAAUUGAAAAUGCCACUGCUAGGGGGGGGAA_CCGGAGGGGG_AGGGGGCAGCAGAAAGCAACCGAAAAAAGGAAAAUCGGAAGGG .(((((......((((((.(((((.......))))).)))....((((.((.((......((....))......)).))..)))))))...)))))((((...........))))..... (-19.11 = -19.30 + 0.19)


| Location | 19,361,686 – 19,361,806 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361686 120 - 23771897 CCCUUCCGAUUUUCCUUUUUUCGGUUGCUUUCUGCUGCCUCCCCCUCCCCCCUGGCUACACCCACUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAAC ......................(((.((.....)).)))...............((((((.((((.....))))....................((((.......))))..))))))... ( -20.70) >DroSec_CAF1 11400 119 - 1 CCCUUCCGAUUUUCCUUUUUUCGGUUGCUUUCUGCCGCCCCCU-CCCCCUCCGGUUUCCCCACCCUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAAC .....((((...........))))((((....((((((.....-....((..(((......)))..))..))))))...................((((((........)))))))))). ( -17.90) >DroSim_CAF1 11694 120 - 1 CCCUUCCGAUUUUCCUUUUUUCGGUUGCUUUCUGCCGCCCCCUUCCCCCUCCGGCUUCCCCACCCUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAAC .....((((...........))))(((((...(((((((.............)))....((((.......))))..............................))))......))))). ( -19.02) >DroYak_CAF1 15711 105 - 1 CCUUUCCGAUUUUCCUUUUUUCGGUUGGUUUCUGCUGCCCCCCA---------------GCCCACUACCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAAC ((...((((...........))))..))....((((((......---------------((((((.....)))((((.................))))......))).....)))))).. ( -20.93) >consensus CCCUUCCGAUUUUCCUUUUUUCGGUUGCUUUCUGCCGCCCCCC_CCCCCUCCGG_UUCCCCACACUAGCAGUGGCAUUUUCAAUUUUACUUUAUAUGCAAAUUUGGCAUUUUGUAGCAAC .....((((...........))))((((....((((((................................))))))...................((((((........)))))))))). (-13.57 = -13.57 + 0.00)



| Location | 19,361,806 – 19,361,923 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -30.99 |
| Energy contribution | -30.87 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19361806 117 - 23771897 AAG---CGCAGCCACAGCAAAGGAGGCGAAGGCGAGAGGUUAAAGCGAUUGUGGACUGAAAAAUGCAGGCCGAGUCAAGAACUGCAUUUGUCGGCUUUCGAAAGCGCUCCUCUUAUUUUU ...---(((..((........))..)))((((..(((((....((((.(((.((.((((.((((((((..(.......)..)))))))).))))))..)))...)))))))))..)))). ( -37.00) >DroSec_CAF1 11519 120 - 1 AAAGAAAUCAGCCACAGCAAAGGAGGCGAAGGCGAGAGGUUAAAGCGAUUGUGGACUGGAAAAUGCAGGCCGAGUCAAGAACUGCAUUUGUCGGCUUUCGAAAGAGCUCCUCUUAUUUUU ..........(((...((.......))...)))(((..(((....(((..((.(((....((((((((..(.......)..))))))))))).))..)))....)))..)))........ ( -34.90) >DroSim_CAF1 11814 119 - 1 AAAGAAAGCAGCCACAGCAAAGGAGGCGAAGGCGAGAGGUUAAAGCGAUUGUGGACUGGAAAAUGCAGGCCGAGUCAAGAACUGCAUUUGUCGGCUUUCGAAAGCGCU-CUCUUAUUUUU ..........(((...((.......))...)))((((((((....(((..((.(((....((((((((..(.......)..))))))))))).))..)))..))).))-)))........ ( -35.50) >DroYak_CAF1 15816 114 - 1 AAA---C---GCCACUGCAAAGGAGGGGAAGCCGAGAGGUUAAAGCGAUUGUGGACUGGAAAAUGCAGGCCGAGUCAAGAACUGCAUUUGUCGGCUUUCGAAAGCCCUCCUCUUAUUUUU ...---.---((....))..(((((((..((((....))))....(((..((.(((....((((((((..(.......)..))))))))))).))..)))....)))))))......... ( -39.10) >consensus AAA___AGCAGCCACAGCAAAGGAGGCGAAGGCGAGAGGUUAAAGCGAUUGUGGACUGGAAAAUGCAGGCCGAGUCAAGAACUGCAUUUGUCGGCUUUCGAAAGCGCUCCUCUUAUUUUU ..........((....)).......((....))((((((......(((..((.(((....((((((((..(.......)..))))))))))).))..)))........))))))...... (-30.99 = -30.87 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:27 2006