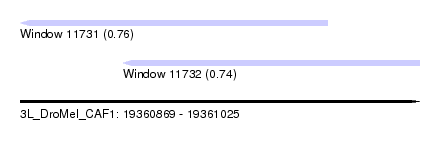
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,360,869 – 19,361,025 |
| Length | 156 |
| Max. P | 0.760865 |

| Location | 19,360,869 – 19,360,989 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -17.72 |
| Energy contribution | -19.16 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
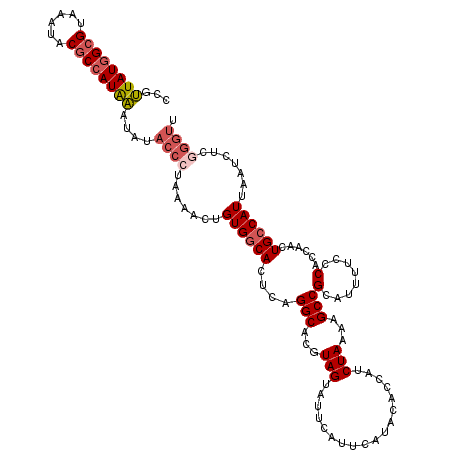
>3L_DroMel_CAF1 19360869 120 - 23771897 CCGUUAUGGCGUAAAUACGCCAUAAAUAUACCCUAAAACUGUGGCACUGUGGCACGUAGUAUUCAUUCAUACACCAUCUAAAAGCCGCAUUUUCCCACCAACUGCCAUUAAUCUCGGGUU ...(((((((((....)))))))))....((((.......((((((.((((((...(((..................)))...)))))).............)))))).......)))). ( -31.75) >DroSec_CAF1 10670 113 - 1 CCGUUAUGGCGUAAAUACACCAUAAAUAUACCCUAAAACUGUGGCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUAAAAGCCGCAUUUUCCCACCAACUGCCAUUAAUC------- ...((((((.((....)).))))))..............((((((....(((...((.((((......))))))..)))....))))))........................------- ( -17.00) >DroSim_CAF1 10922 120 - 1 CCGUUAUGGCGUAAAUACGCCAUAAAUAUACCCUAAAACUGUGGCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUAAAAGCCGCAUUUUCCCACCAACUGCCAUUAAUCUCGGGUU ...(((((((((....)))))))))....((((.......((((((....(((...(((..................)))...)))(........)......)))))).......)))). ( -27.51) >DroYak_CAF1 14813 117 - 1 ---CUAUUGCGAAAAUACGCCAUGGAUAUACCAUAACACUGUGCCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUAAAAGCCGCAUUUUCCCAUCAACUGCCAUUAAUCUCGGGUA ---....((((....(((...((((.....))))......(((((.....))))))))((((......)))).............))))....(((...................))).. ( -20.41) >consensus CCGUUAUGGCGUAAAUACGCCAUAAAUAUACCCUAAAACUGUGGCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUAAAAGCCGCAUUUUCCCACCAACUGCCAUUAAUCUCGGGUU ...((((((((......))))))))....((((.......((((((....(((...(((..................)))...)))(........)......)))))).......)))). (-17.72 = -19.16 + 1.44)

| Location | 19,360,909 – 19,361,025 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.64 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
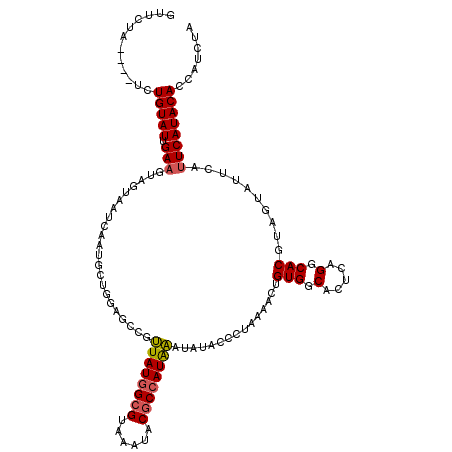
>3L_DroMel_CAF1 19360909 116 - 23771897 CUUCUA----UCUGUAUUGAUGAAGUAAUCAAUGCAGGAGCCGUUAUGGCGUAAAUACGCCAUAAAUAUACCCUAAAACUGUGGCACUGUGGCACGUAGUAUUCAUUCAUACACCAUCUA ......----(((((((((((......))))))))))).(((((((((((((....))))))))).....(((.......).)).....)))).....((((......))))........ ( -32.20) >DroSec_CAF1 10703 111 - 1 -----A----UCUGUAUUGAAGUAGUAAUCAAUGGUGGAGCCGUUAUGGCGUAAAUACACCAUAAAUAUACCCUAAAACUGUGGCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUA -----.----.....(((((........)))))(((((.(((.((((((.((....)).)))))).....(((.......).))......))).....((((......)))).))))).. ( -21.70) >DroSim_CAF1 10962 116 - 1 GUUCUA----UCUGUAUUGAAGUAGUAAUCAAUGCUGCAGCCGUUAUGGCGUAAAUACGCCAUAAAUAUACCCUAAAACUGUGGCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUA ......----...........(((..(((.((((((((.(((.(((((((((....))))))))).....(((.......).))......)))..)))))))).)))..)))........ ( -28.30) >DroYak_CAF1 14853 102 - 1 GUGCUAUGUGUCUGUAUUGAAGUAG------------------CUAUUGCGAAAAUACGCCAUGGAUAUACCAUAACACUGUGCCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUA ((((((((((((((...((..(((.------------------((((.(((......))).))))...)))((((....)))).))..)))))))))))))).................. ( -27.80) >consensus GUUCUA____UCUGUAUUGAAGUAGUAAUCAAUGCUGGAGCCGUUAUGGCGUAAAUACGCCAUAAAUAUACCCUAAAACUGUGGCACUCAGGCACGUAGUAUUCAUUCAUACACCAUCUA ............(((((.(((......................((((((((......))))))))...............(((.(.....).)))..........))))))))....... (-16.04 = -16.22 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:17 2006