
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,356,504 – 19,356,692 |
| Length | 188 |
| Max. P | 0.891769 |

| Location | 19,356,504 – 19,356,616 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -18.91 |
| Energy contribution | -21.35 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19356504 112 - 23771897 UUUUCCUGGCCAUCCUUUUGGCCCGAGGGACGACU--------AUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCUUAUCUCUGAUUAUUAUCCUUCUAGCUCAGGAGCUCAGGAA ..((((((((((......)))))(((((((((...--------...))....((....))...)))))))((((((...........(((....)))..........))))))..))))) ( -30.20) >DroSec_CAF1 6492 120 - 1 UUUUCCUGACCAUCCUUUUGGCCCCAGGGACGACUGGGGCUGCAUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCUUAUCUCUGAUUAUUAUCCUUCUAGCUCAGAAGCUCAGGAA ..(((((((..........((((((((......))))))))((..((((..................)))).............((((((((.........))).))))).))))))))) ( -32.67) >DroSim_CAF1 6559 120 - 1 UUUUCCUGGCCAUCCUUUUGGCCCCAGGGACGACUGGGGCUGCAUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCUUAUCUCUGAUUAUUAACCUUCUAGCUCAGGAGCUCAGGAA ..((((((((..((((...((((((((......))))))))((..((((..................))))......((........))..............))..)))))).)))))) ( -33.77) >DroYak_CAF1 10342 112 - 1 UUUUCCUGGCCAUCCUUUUGGCUUAAGGGGCGACUGGAGCUGCAUCCGAUUAUCCUACGACUUUCCUUCGUCUCCUGCCUUAUCUCUGAUUAUUAUCCUUCUAGCU--------CAGGAA ..((((((((((......))))....((((((..((((......))))..........(((........)))...)))))).........................--------)))))) ( -25.10) >consensus UUUUCCUGGCCAUCCUUUUGGCCCCAGGGACGACUGGGGCUGCAUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCUUAUCUCUGAUUAUUAUCCUUCUAGCUCAGGAGCUCAGGAA ..((((((((((......))))..((((((..(..(((((((.....((...((....))...))...)))))))....)..))))))..........................)))))) (-18.91 = -21.35 + 2.44)

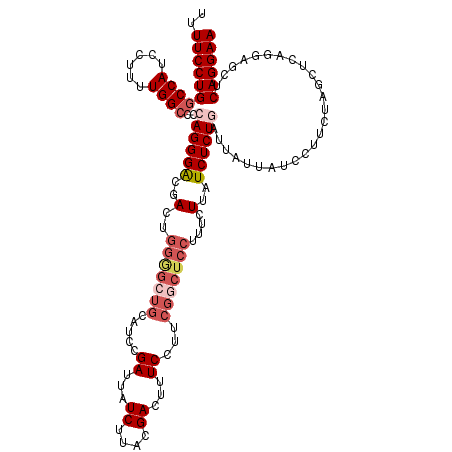

| Location | 19,356,544 – 19,356,656 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -29.84 |
| Energy contribution | -30.90 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19356544 112 - 23771897 GCAUGUGGCUAAGGAAUGGCUUACAAGCCGGUUCCUUGUUUUUUCCUGGCCAUCCUUUUGGCCCGAGGGACGACU--------AUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCU ....(..((((((((((((((....)))))((((((((.........(((((......)))))))))))))....--------...................)))))).)))..)..... ( -34.10) >DroSec_CAF1 6532 120 - 1 GCAUGUGGCUAAGGAAUGGCUUACAAGCCGCUUCCCUGUUUUUUCCUGACCAUCCUUUUGGCCCCAGGGACGACUGGGGCUGCAUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCU ....(..(((((((((.((((....))))((..((((..((.((((((.(((......)))...)))))).))..))))..))...................)))))).)))..)..... ( -36.20) >DroSim_CAF1 6599 120 - 1 GCAUGUGGCUAAGGAAUGGCUUACAAGCCGCUUCCCUGUUUUUUCCUGGCCAUCCUUUUGGCCCCAGGGACGACUGGGGCUGCAUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCU ....(..(((((((((.((((....))))((..((((..((.((((((((((......)))))..))))).))..))))..))...................)))))).)))..)..... ( -40.20) >DroYak_CAF1 10374 120 - 1 GCAUGUGGCUGAGGAAUGGCUUACAAGCCGUUUCUCUGUUUUUUCCUGGCCAUCCUUUUGGCUUAAGGGGCGACUGGAGCUGCAUCCGAUUAUCCUACGACUUUCCUUCGUCUCCUGCCU (((((..((((((((((((((....))))))))))).(((..((((((((((......)))))..))))).)))...)))..))..............(((........)))...))).. ( -39.10) >consensus GCAUGUGGCUAAGGAAUGGCUUACAAGCCGCUUCCCUGUUUUUUCCUGGCCAUCCUUUUGGCCCCAGGGACGACUGGGGCUGCAUCCGAUUAUCUUACGACUUUCCUUCGGCUCCUUUCU ....(..(((((((((.((((....))))((..(((((.((.((((((((((......)))))..))))).)).)))))..))...................)))))).)))..)..... (-29.84 = -30.90 + 1.06)

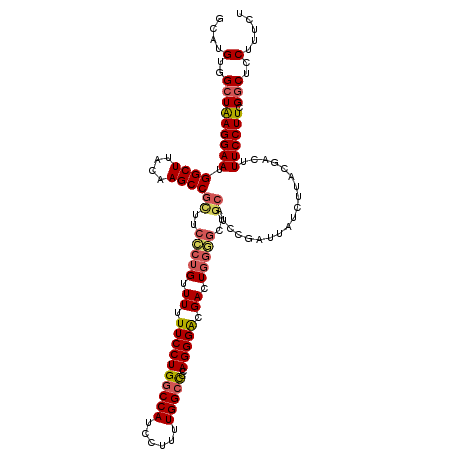
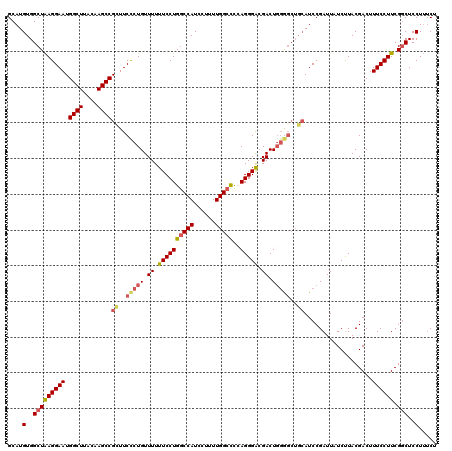
| Location | 19,356,581 – 19,356,692 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -31.47 |
| Energy contribution | -33.03 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19356581 111 + 23771897 -----AGUCGUCCCUCGGGCCAAAAGGAUGGCCAGGAAAAAACAAGGAACCGGCUUGUAAGCCAUUCCUUAGCCACAUGCCUACAAACAGGA----AAAACCAAGGCCAGAAAGCAGUCC -----....((...((.((((...(((.((((.(((((.......(....)((((....)))).)))))..))))....))).......((.----....))..)))).))..))..... ( -28.70) >DroSec_CAF1 6572 120 + 1 GCCCCAGUCGUCCCUGGGGCCAAAAGGAUGGUCAGGAAAAAACAGGGAAGCGGCUUGUAAGCCAUUCCUUAGCCACAUGCCUGCAAACAGGGCCAACCAACCAAGGCCAGAAAGCGGGCC (((((((......)))))))........((((..(.......)((((((..((((....)))).)))))).))))...((((((......((((..........)))).....)))))). ( -45.10) >DroSim_CAF1 6639 120 + 1 GCCCCAGUCGUCCCUGGGGCCAAAAGGAUGGCCAGGAAAAAACAGGGAAGCGGCUUGUAAGCCAUUCCUUAGCCACAUGCCUGCAAACAGGGCCAACCAACCAAGGCCAGAAAGCGGGCC (((((((......)))))))........((((..(.......)((((((..((((....)))).)))))).))))...((((((......((((..........)))).....)))))). ( -47.10) >consensus GCCCCAGUCGUCCCUGGGGCCAAAAGGAUGGCCAGGAAAAAACAGGGAAGCGGCUUGUAAGCCAUUCCUUAGCCACAUGCCUGCAAACAGGGCCAACCAACCAAGGCCAGAAAGCGGGCC (((((((......)))))))........(((((.((.......((((((..((((....)))).)))))).........((((....)))).........))..)))))........... (-31.47 = -33.03 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:10 2006