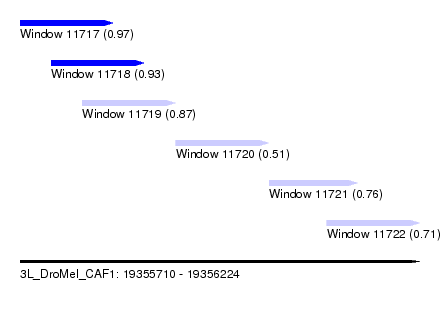
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,355,710 – 19,356,224 |
| Length | 514 |
| Max. P | 0.969258 |

| Location | 19,355,710 – 19,355,830 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.27 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19355710 120 + 23771897 UCCUUCCCCCUUUCAGUUGCCACUGGUCUACUGUUAUUGUUGCUGUGGCUUUCGCUUUGGCAUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGA (((...............(((((.(((..((.......)).)))))))).........(((((((((....))))))))).(((((((((((((.....)))))).)))))))....))) ( -45.00) >DroSec_CAF1 5739 90 + 1 UCCUUCCUCCUUUCAGUUGCUCCUGGUCUAC------------------------------UUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGA ....(((.................((.(...------------------------------.(((((....))))).).))(((((((((((((.....)))))).)))))))....))) ( -30.00) >DroSim_CAF1 5777 114 + 1 UCCUUCCUCCUUUCAGUUGCACCUGGUCUACUGUUAUUGUUGCUGUGGCUU------UGGCUUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGA ....(((...........(((((.(((..((.......)).)))..((((.------.(((....)))..)))))))))..(((((((((((((.....)))))).)))))))....))) ( -37.60) >DroYak_CAF1 9563 107 + 1 UCUUUCC-CCUUUUAGUUGCUCCCGAGGUAGUGUUAUUGUUGCUGUGGCUU------------UGGCUUUGGUUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGAA .......-..........(((.(((((((...(((((.......)))))..------------..)))))))..)))....(((((((((((((.....)))))).)))))))....... ( -29.70) >consensus UCCUUCCUCCUUUCAGUUGCUCCUGGUCUACUGUUAUUGUUGCUGUGGCUU__________UUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGA ............(((((((...((((...............((....)).............))))...))))))).....(((((((((((((.....)))))).)))))))....... (-22.83 = -22.27 + -0.56)


| Location | 19,355,750 – 19,355,870 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.96 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -30.02 |
| Energy contribution | -31.90 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19355750 120 + 23771897 UGCUGUGGCUUUCGCUUUGGCAUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAAGGGGAUACACAAAGGAUUUGAGC ...((((...(((.(((((((((((((....))))))))).(((((((((((((.....)))))).))))))).....................)))).)))..))))............ ( -43.60) >DroSec_CAF1 5770 99 + 1 ---------------------UUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAGGGGGAUACACAAAGGAUUUGAGC ---------------------..((((....))))(((((((((((((((((((.....)))))).))))))....((.....)).............)))))))............... ( -34.30) >DroSim_CAF1 5817 114 + 1 UGCUGUGGCUU------UGGCUUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAGGGGGAUACACAAAGGAUUUGAGC .(((..((((.------.(((....)))..)))).(((((((((((((((((((.....)))))).))))))....((.....)).............)))))))............))) ( -39.10) >DroYak_CAF1 9602 108 + 1 UGCUGUGGCUU------------UGGCUUUGGUUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGAAAAACAAAAGGCUUAAAAGGGGAUACACAAAGGAUUUGAGC .......((((------------...(((((....(((((((((((((((((((.....)))))).))))))..................((....))))))))).))))).....)))) ( -32.30) >consensus UGCUGUGGCUU__________UUCGGCUUCGGCUGGUGUCCUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAAGGGGAUACACAAAGGAUUUGAGC .......................((((....))))(((((((((((((((((((.....)))))).))))))....((.....)).............)))))))............... (-30.02 = -31.90 + 1.88)


| Location | 19,355,790 – 19,355,910 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.03 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -33.80 |
| Energy contribution | -35.05 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19355790 120 + 23771897 CUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAAGGGGAUACACAAAGGAUUUGAGCAAUCAAGUGCGCUUUUGAGCACGCAGACCGAUAACAGUUU .(((((((((((((.....)))))).))))))).......((((.....(((((((((.(......)...(.((((((....)))))).).))))))))).((.....))......)))) ( -35.20) >DroSec_CAF1 5789 120 + 1 CUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAGGGGGAUACACAAAGGAUUUGAGCAAUCAAGUGCGCUUUUGAGCACGCAGACCGAUAACAGUUU .(((((((((((((.....)))))).))))))).......((((.....(((((((((.(......)...(.((((((....)))))).).))))))))).((.....))......)))) ( -34.90) >DroSim_CAF1 5851 120 + 1 CUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAGGGGGAUACACAAAGGAUUUGAGCAAUCAAGUGCGCUUUUGAGCACGCAGACCGAUAACAGUUU .(((((((((((((.....)))))).))))))).......((((.....(((((((((.(......)...(.((((((....)))))).).))))))))).((.....))......)))) ( -34.90) >DroYak_CAF1 9630 120 + 1 CUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGAAAAACAAAAGGCUUAAAAGGGGAUACACAAAGGAUUUGAGCAAUCAAGUGCGCUUUUGAGCACGCAGACCGAUAACAGUUU .(((((((((((((.....)))))).))))))).......((((.....(((((((((.(......)...(.((((((....)))))).).))))))))).((.....))......)))) ( -35.20) >consensus CUGCGCUGAAGUGUUUACAACACUUCCGGCGCAUAAUGGAAAACAAAAGGCUUAAAAGGGGAUACACAAAGGAUUUGAGCAAUCAAGUGCGCUUUUGAGCACGCAGACCGAUAACAGUUU .(((((((((((((.....)))))).))))))).......((((.....(((((((((.(......)...(.((((((....)))))).).))))))))).((.....))......)))) (-33.80 = -35.05 + 1.25)


| Location | 19,355,910 – 19,356,030 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.91 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -28.96 |
| Energy contribution | -28.77 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.99 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19355910 120 + 23771897 UAUUCACGCUCGCUUUUUCCAGCCUCUUAUUUUCCUGGCACCUGCCAUUAGGUUGAGGGCUAAGAAGAAAAUGUGCUUUCGCCCUCAGUUUGUUUCAAAGUCACAAACAAUGCUGGCUUU ..................(((((..........((((((....)))...))).(((((((...((((........)))).)))))))((((((.........))))))...))))).... ( -30.10) >DroSec_CAF1 5909 119 + 1 UAUUCACGCUCGCUUUUUCCAGCCUCUUAUUUUCCUGGCACCUGCCAUUAGGUUGAGGGCUAAGAAGAAAAUGUGCUUUCGCCCUCAGUUUAUUUCAAAGUCAUA-ACAAUGCUGGCUUU ..................(((((............((((....)))).((..((((((((...((((........)))).))))))))..)).............-.....))))).... ( -29.30) >DroSim_CAF1 5971 119 + 1 UAUUCACGCUCGCUUUUUCCAGCCUCUUAUUUUCCUGGCACCUGCCAUUAGGUUGAGGGCUAAGAAGAAAAUGUGCUUUCGCCCUCAGUUUGUUUCAAAGUCAUA-ACAAUGCUGGCUUU ..................(((((............((((....)))).((..((((((((...((((........)))).))))))))..)).............-.....))))).... ( -29.00) >DroYak_CAF1 9750 119 + 1 UAUUCACGCUCGCUUUUUCCAGCCUCUUAUUUUCCUGGCACCUGCCAUUACGUUGAGGGCUAAGAAGAAAAUGUGCUUUCGCCCUCAGUUUGUUUCAAAGUCAUA-ACAAUGCUGGGUUU ....((((....((((((..((((.((((......((((....))))......))))))))))))))....))))........(((((((((((..........)-)))).))))))... ( -29.00) >consensus UAUUCACGCUCGCUUUUUCCAGCCUCUUAUUUUCCUGGCACCUGCCAUUAGGUUGAGGGCUAAGAAGAAAAUGUGCUUUCGCCCUCAGUUUGUUUCAAAGUCAUA_ACAAUGCUGGCUUU ..................(((((............((((....)))).((..((((((((...((((........)))).))))))))..))...................))))).... (-28.96 = -28.77 + -0.19)



| Location | 19,356,030 – 19,356,144 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19356030 114 + 23771897 UCAAUAUGCGCCCAAAGGAACCCACACGUAUGCGCCAUAAAGUCCUCCCAUGUGGCAGGAAAAA------AGGGAAAAAUAAGUAAGGAAACGGGGCAUAAAACUUCAAGUAGUUGGGUC .........((((((.............(((((.((......((((.((....)).))))....------................(....))).))))).............)))))). ( -27.97) >DroSec_CAF1 6028 110 + 1 UCAAUAUGCGCCCAAAGGAACCCACAC----GCGCCAUAAAGUCCUCCCAUGAGGCAGGAAAAA------AGGGAAAAAUAAGCAAGGAAACGGGGCAUAAAACUUCAAGUAGUUAGGUC ....(((((.(((..............----((.((......((((.((....)).))))....------..))........))..(....))))))))).((((......))))..... ( -25.00) >DroSim_CAF1 6090 114 + 1 UCAAUAUGCGCCCAAAGGAACCCACACGUAUGCGCCAUAAAGUCCUCCCAUGGGGCAGGAAAAA------AGGGAAAAAUAAGCAAGGAAACGGGGCAUAAAACUUCAAGUAGUUAGGUC ....(((((.(((.......(((...((....))........(((((((...))).))))....------.)))............(....))))))))).((((......))))..... ( -28.00) >DroYak_CAF1 9869 120 + 1 UCAAUAUGCGCCCAAAGGAACCCACACGUAUGCGCCAUAAAGUCCUCCCAUAAGGCAGGAAAAAAGGAAAAGGGAAAAAUAAGUAAGGAAACGGGGCAUAAAACUUCAAGUAGUUGGGUC ....(((((.(((.......(((...((....))((......((((.((....)).)))).....))....)))............(....))))))))).((((......))))..... ( -28.00) >consensus UCAAUAUGCGCCCAAAGGAACCCACACGUAUGCGCCAUAAAGUCCUCCCAUGAGGCAGGAAAAA______AGGGAAAAAUAAGCAAGGAAACGGGGCAUAAAACUUCAAGUAGUUAGGUC ....(((((.(((.......(((...((....))........((((.((....)).))))...........)))............(....))))))))).((((......))))..... (-25.25 = -25.50 + 0.25)



| Location | 19,356,104 – 19,356,224 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -31.33 |
| Energy contribution | -31.32 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19356104 120 + 23771897 AAGUAAGGAAACGGGGCAUAAAACUUCAAGUAGUUGGGUCCAAAUGUCUGCACGGCUUUAGCCAGCAACAACAAUGCGGCCCGAAAGGGUGGACACAGAAAAAAAGGAUUCCGAGGGGUC ......(....).((((....((((......))))..))))...(((((....(((....))).(((.......))).((((....)))))))))...........(((((....))))) ( -33.30) >DroSec_CAF1 6098 120 + 1 AAGCAAGGAAACGGGGCAUAAAACUUCAAGUAGUUAGGUCCAAAUGUCUGCACGGCUUUAGCCAGCAACAACAAUGCGGCCCGAAAGGGUGGACACAGAAAAUAAGGAUUCCGAGGGGUU ..((..(....)...))......((((..........((((...(((.(((..(((....))).))))))........((((....))))))))...........((...)))))).... ( -32.90) >DroSim_CAF1 6164 120 + 1 AAGCAAGGAAACGGGGCAUAAAACUUCAAGUAGUUAGGUCCAAAUGUCUGCACGGCUUUAGCCAGCAACAACAAUGCGGCCCGAAAGGGUGGACACAGAAAAAAAGGAUUCCGAGGGGUC ..((..(....)...))....((((......))))..((((...(((.(((..(((....))).))))))........((((....))))))))............(((((....))))) ( -33.30) >DroYak_CAF1 9949 119 + 1 AAGUAAGGAAACGGGGCAUAAAACUUCAAGUAGUUGGGUCCAAACGUCUGCACGGCUUUAGCCAGCAACAACAAUACGGCCCGGAAGGGUGGACACAGAAAA-AAGAAUUCCGAGCGGUC ..((..((((...((((....((((......))))..))))....((((((..(((....))).))............((((....))))))))........-.....))))..)).... ( -33.60) >consensus AAGCAAGGAAACGGGGCAUAAAACUUCAAGUAGUUAGGUCCAAAUGUCUGCACGGCUUUAGCCAGCAACAACAAUGCGGCCCGAAAGGGUGGACACAGAAAAAAAGGAUUCCGAGGGGUC ......((((...((((....((((......))))..))))....((((((..(((....))).))............((((....))))))))..............))))........ (-31.33 = -31.32 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:07 2006