
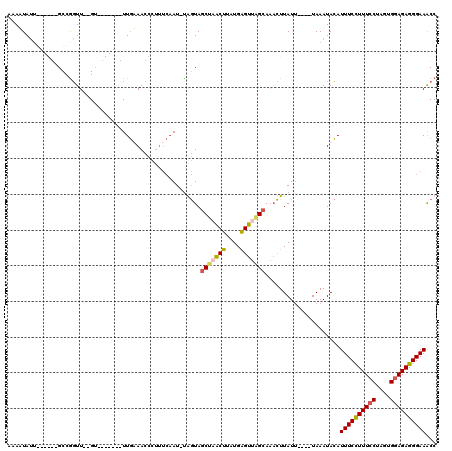
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,351,766 – 19,351,950 |
| Length | 184 |
| Max. P | 0.999938 |

| Location | 19,351,766 – 19,351,870 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.85 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.58 |
| SVM decision value | 4.68 |
| SVM RNA-class probability | 0.999938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19351766 104 + 23771897 AAAAUAUU------GCCGGUU--GU-------UUGUAACCCUUUCAAU-UAGUAGCUUGCUUUUGAGUUUGCAAACUUAUAUACAUAAAUACAUUUCCUUUCCUAUUGGAGAGGGAAACC ........------...((((--((-------((((((....(((((.-.(((.....))).))))).))))))))..................(((((((((....))))))))))))) ( -25.80) >DroSec_CAF1 1920 113 + 1 AUAUUAUUGUAGGUGCCGGUU--GCUAUUAACUUGAAACCCUUUCAAU-UGGUAGCUAACUUAUGAGUUAGCAAACUUAUU----UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACC .((((...(((((((((((((--(....................))))-)))).(((((((....)))))))..)))))).----..))))..((((((((((....))))))))))... ( -32.65) >DroSim_CAF1 1946 113 + 1 AUAUUAUUGUAGGUGCCAGUU--GAUUUUAACUUGAAACCCUUUCAAU-UGGUAGCUAACUUAUGAGUUAGCAAACUUAUU----UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACC .((((...(((((((((((((--((..................)))))-)))).(((((((....)))))))..)))))).----..))))..((((((((((....))))))))))... ( -36.57) >DroYak_CAF1 5293 93 + 1 AAAAUAGG------GCCGUUUUAGU-------UUUUAA--------AUACAAUACCAUGCUUGUGGGUUUGCA--UUUAUU----UAAAUAGAUUUCUUUUCCCUUUGCAGAGGGAAGCC .......(------((.......((-------((.(((--------((.(((..((((....))))..))).)--))))..----.)))).........(((((((....)))))))))) ( -20.10) >consensus AAAAUAUU______GCCGGUU__GU_______UUGAAACCCUUUCAAU_UAGUAGCUAACUUAUGAGUUAGCAAACUUAUU____UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACC ......................................................(((((((....))))))).....................((((((((((....))))))))))... (-16.83 = -17.20 + 0.37)


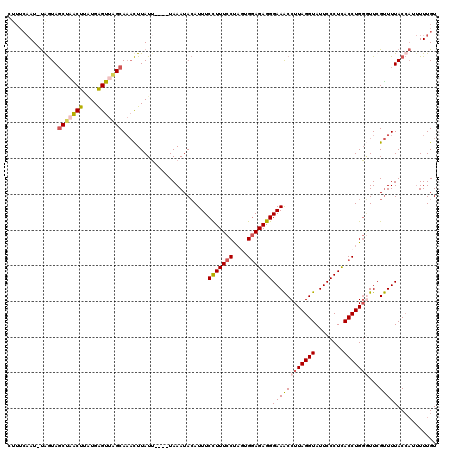
| Location | 19,351,791 – 19,351,910 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19351791 119 + 23771897 CUUUCAAU-UAGUAGCUUGCUUUUGAGUUUGCAAACUUAUAUACAUAAAUACAUUUCCUUUCCUAUUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGUAUUCGUUUUACCAUUUUUGU ........-..(((((..(((....)))..))............(..((((((.......(((....)))((((((((((...))).)))))))...))))))..)..)))......... ( -23.90) >DroSec_CAF1 1958 115 + 1 CUUUCAAU-UGGUAGCUAACUUAUGAGUUAGCAAACUUAUU----UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGGGUUCGUUUUACCAUUUUUGU ....(((.-((((((((((((....)))))))((((.....----........((((((((((....))))))))))((((.((((........))))))))..)))))))))...))). ( -36.10) >DroSim_CAF1 1984 115 + 1 CUUUCAAU-UGGUAGCUAACUUAUGAGUUAGCAAACUUAUU----UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGGGUUCGUUUUACCAUUUUUGU ....(((.-((((((((((((....)))))))((((.....----........((((((((((....))))))))))((((.((((........))))))))..)))))))))...))). ( -36.10) >DroYak_CAF1 5318 108 + 1 ------AUACAAUACCAUGCUUGUGGGUUUGCA--UUUAUU----UAAAUAGAUUUCUUUUCCCUUUGCAGAGGGAAGCCUUAGGUAUUCCCUUACCUGGCUUCGCUUUACAUUCUUUGU ------..((((......((....(((.....(--(((((.----...)))))).......)))...))((((.((((((..(((((......))))))))))).)))).......)))) ( -27.00) >consensus CUUUCAAU_UAGUAGCUAACUUAUGAGUUAGCAAACUUAUU____UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGGGUUCGUUUUACCAUUUUUGU ..............(((((((....)))))))........................(((((((....)))))))((((((((((((........)))))))...)))))........... (-21.38 = -22.12 + 0.75)


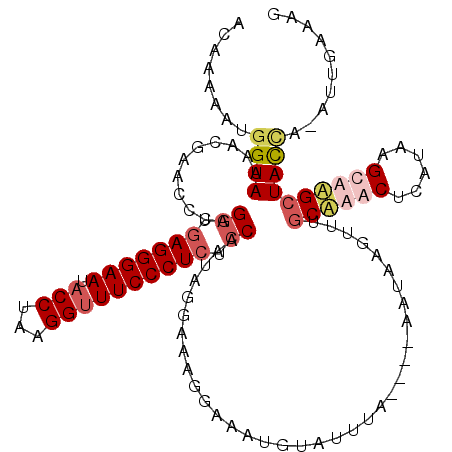
| Location | 19,351,791 – 19,351,910 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -17.51 |
| Energy contribution | -19.45 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19351791 119 - 23771897 ACAAAAAUGGUAAAACGAAUACAGGUGAGGGAAUACCUAAGGUUUCCCUCUCCAAUAGGAAAGGAAAUGUAUUUAUGUAUAUAAGUUUGCAAACUCAAAAGCAAGCUACUA-AUUGAAAG .......(((((....(((((((...(((((((.(((...))))))))))(((....))).......)))))))..........((((((..........)))))))))))-........ ( -28.80) >DroSec_CAF1 1958 115 - 1 ACAAAAAUGGUAAAACGAACCCAGGUGAGGGAAUACCUAAGGUUUCCCUCUCCACUAGGAAAGGAAAUGUAUUUA----AAUAAGUUUGCUAACUCAUAAGUUAGCUACCA-AUUGAAAG .......(((((((((.......((.(((((((.(((...)))))))))).)).((.....))............----.....))))(((((((....))))))))))))-........ ( -30.50) >DroSim_CAF1 1984 115 - 1 ACAAAAAUGGUAAAACGAACCCAGGUGAGGGAAUACCUAAGGUUUCCCUCUCCACUAGGAAAGGAAAUGUAUUUA----AAUAAGUUUGCUAACUCAUAAGUUAGCUACCA-AUUGAAAG .......(((((((((.......((.(((((((.(((...)))))))))).)).((.....))............----.....))))(((((((....))))))))))))-........ ( -30.50) >DroYak_CAF1 5318 108 - 1 ACAAAGAAUGUAAAGCGAAGCCAGGUAAGGGAAUACCUAAGGCUUCCCUCUGCAAAGGGAAAAGAAAUCUAUUUA----AAUAAA--UGCAAACCCACAAGCAUGGUAUUGUAU------ ........((((.((.(((((((((((......)))))..)))))).)).)))).....................----.....(--(((((..(((......)))..))))))------ ( -24.70) >consensus ACAAAAAUGGUAAAACGAACCCAGGUGAGGGAAUACCUAAGGUUUCCCUCUCCAAUAGGAAAGGAAAUGUAUUUA____AAUAAGUUUGCAAACUCAUAAGCAAGCUACCA_AUUGAAAG ........((((...........((.(((((((.(((...)))))))))).))...................................((((((......)))))))))).......... (-17.51 = -19.45 + 1.94)


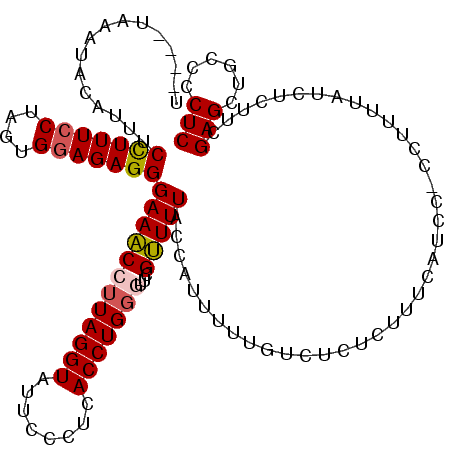
| Location | 19,351,830 – 19,351,950 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19351830 120 + 23771897 AUACAUAAAUACAUUUCCUUUCCUAUUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGUAUUCGUUUUACCAUUUUUGUCUCUGUUUUAUCCUCCUUUUAUCUCUUCGAGCUGCCCCUC ....(..((((((.......(((....)))((((((((((...))).)))))))...))))))..)..........................................(((......))) ( -19.10) >DroSec_CAF1 1997 115 + 1 U----UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGGGUUCGUUUUACCAUUUUUGUCUCUCUUUCAUCC-CCUUUUAUCUCUUCGAGCUGCCCCUC .----........((((((((((....))))))))))......((((..(((......))).......))))......((..(((........-..............)))..))..... ( -21.65) >DroSim_CAF1 2023 115 + 1 U----UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGGGUUCGUUUUACCAUUUUUGUCUCUCUUUCAUCC-CCUUUUAUCUCUUCGAGCUGCCGCUC .----........((((((((((....))))))))))......((((..(((......))).......)))).....................-..............((((....)))) ( -25.40) >DroYak_CAF1 5350 116 + 1 U----UAAAUAGAUUUCUUUUCCCUUUGCAGAGGGAAGCCUUAGGUAUUCCCUUACCUGGCUUCGCUUUACAUUCUUUGUCUCUGUUUUAUCCUUCUUUUAUCUCUUCGAGCUGCCCCUC (----..((.((((.............((((((.((((((..(((((......))))))))))).....(((.....)))))))))..............)))).))..).......... ( -24.23) >consensus U____UAAAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACCUGGGUUCGUUUUACCAUUUUUGUCUCUCUUUCAUCC_CCUUUUAUCUCUUCGAGCUGCCCCUC ................(((((((....)))))))((((((((((((........)))))))...))))).......................................(((......))) (-16.31 = -16.75 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:56 2006