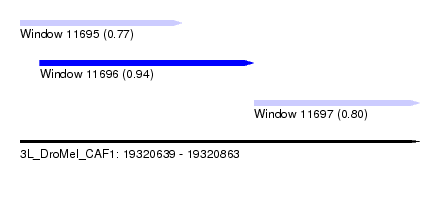
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,320,639 – 19,320,863 |
| Length | 224 |
| Max. P | 0.937581 |

| Location | 19,320,639 – 19,320,730 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.79 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -26.54 |
| Energy contribution | -27.07 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19320639 91 + 23771897 --------------------C---------UCAACUCACCCGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUC --------------------.---------.......((((((....)))....((((((((...(((((.((((((..(((....))))))))).))))))))))))).......))). ( -27.40) >DroVir_CAF1 100446 102 + 1 ------------------GCCUCAAACGUUUUCACUCACCCGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUC ------------------((((...(((((...((......))...)))))...((((((((...(((((.((((((((........)))))))).)))))))))))))......)))). ( -32.20) >DroPse_CAF1 90592 98 + 1 --------------------CCCUGUCG--UCGACUUACCCGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUC --------------------.((((.((--(.(((......)))...)))....((((((((...(((((.((((((..(((....))))))))).))))))))))))).....)))).. ( -32.50) >DroGri_CAF1 125748 104 + 1 ----------------CUGCCUCAAACGUUUCCACUCACCCGUCCAAGCGUAUUGUUAUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUC ----------------..((((...((((((..((......))..))))))...((((((((...(((((.((((((((........)))))))).)))))))))))))......)))). ( -33.90) >DroWil_CAF1 71709 98 + 1 --------------------CCCUUUCG--UUUACUCACCCUGCCCAAUGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUC --------------------........--.........((((.....((....((((((((...(((((.((((((((........)))))))).)))))))))))))))...)))).. ( -28.50) >DroMoj_CAF1 114475 120 + 1 UGGCAUGGUAUGGUAGUCAUCUCAAACGUUUUCACUCACCCGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUC .(((.((((.((((........((((((((...((......))...))))).))).((((((...(((((.((((((((........)))))))).))))))))))))))).)))).))) ( -36.00) >consensus ____________________CUCAAACG__UCCACUCACCCGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUC .....................................((((((....)))....((((((((...(((((.((((((((........)))))))).))))))))))))).......))). (-26.54 = -27.07 + 0.53)

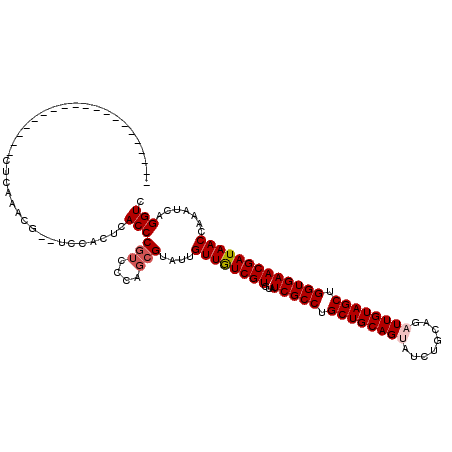
| Location | 19,320,650 – 19,320,770 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -36.85 |
| Energy contribution | -36.88 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19320650 120 + 23771897 CGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAUGUCGUAUUC ((.(...((((((..((..((..((((((...((((((...))))))...))..(((((((..(((......)))...)))))))))))..))....))..))))))....).))..... ( -37.80) >DroVir_CAF1 100468 120 + 1 CGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCACGUUGUGUUC .....((((((...((((((((...(((((.((((((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))......))))))..... ( -39.60) >DroGri_CAF1 125772 120 + 1 CGUCCAAGCGUAUUGUUAUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCACGUUGUGUUC (((....(((((((((((((((...(((((.((((((((........)))))))).))))))))))))).........((((..((....))))))....)))))))..)))........ ( -38.60) >DroWil_CAF1 71727 120 + 1 CUGCCCAAUGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAUGUUGUAUUU .(((...((((((.((((((((...(((((.((((((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))..))))))...)))... ( -38.50) >DroMoj_CAF1 114515 120 + 1 CGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCACGUUGUGUUC .....((((((...((((((((...(((((.((((((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))......))))))..... ( -39.60) >DroPer_CAF1 90302 120 + 1 CGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCACGUUGUGUUC .....((((((...((((((((...(((((.((((((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))......))))))..... ( -38.40) >consensus CGUCCCAGCGUAUUGUUGUCGUGUAUCGCCUGCUGCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCACGUUGUGUUC .....((((((...((((((((...(((((.((((((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))......))))))..... (-36.85 = -36.88 + 0.03)

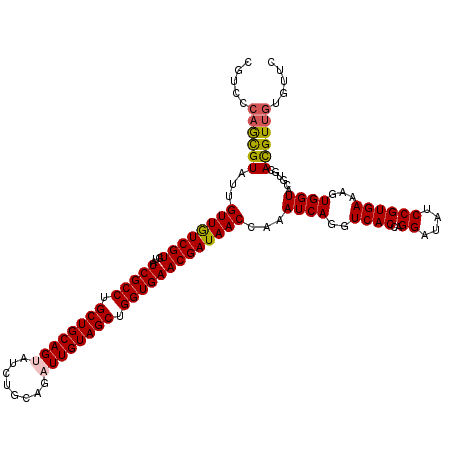
| Location | 19,320,770 – 19,320,863 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.91 |
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -25.79 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19320770 93 + 23771897 GGCGGAAGCGUGUGACGCGGACUCACAAUGCGUCCCAUGCGGCCGCGCCGCGAGAAUAGCACAUCAUCUGGA------AAGGAUGCA----AA-AAAAUCAAGC---- .((((..(((((.((((((.........)))))).)))))..))))((.((.......))....(((((...------..)))))..----..-........))---- ( -30.60) >DroVir_CAF1 100588 97 + 1 GGCGGAAGCGUGUGACGCGGACUCACAAUGCGUCCCAUGCGGCCGCGACGUGAGAAUAAAACAUCAUCUGCG------AAGGAGAAAGACAGA-GAGAUAUACA---- .((((..(((((.((((((.........)))))).)))))..))))..(....)........(((.(((((.------.........).))))-..))).....---- ( -30.70) >DroPse_CAF1 90730 100 + 1 GGCGGAAGCGUGUGACGCGGACUCACAAUGCGUCCCAUGCGGCCGCGACGCGAGAAUAGCACAUCAUCUGCAGAGUAAAAGAAGGCA----AAAAA--UAGAGUCA-- .((((..(((((.((((((.........)))))).)))))..))))(((((.......))........(((.............)))----.....--....))).-- ( -31.52) >DroGri_CAF1 125892 98 + 1 GGCGGAAGCGUGUGACGCGGGCUCACAAUGCGUCCCAUGCGGCCGCGACGUGAGAAUAGCACGUCAUCUGCA------AAGGAGAGA--CAGA-AAGAUAGACAG-AU .((((..(((((.((((((.........)))))).)))))..))))((((((.......)))))).(((((.------.......).--))))-...........-.. ( -36.80) >DroEre_CAF1 153914 92 + 1 GGCGGAAGCGUGUGACGCGGACUCACAAUGCGUCCCAUGCGGCCGCGGCGCGAGAAUAGCACAUCAUCUGGA------AAGG-GGCA----GA-GAAAUCGGGA---- .((((..(((((.((((((.........)))))).)))))..)))).((((.......))......(((...------.)))-.)).----((-....))....---- ( -32.60) >DroPer_CAF1 90422 100 + 1 GGCGGAAGCGUGUGACGCGGACUCACAAUGCGUCCCAUGCGGCCGCGACGCGAGAAUAGCACAUCAUCUGCAGAGUAAAAGAAGGCA----AAAAA--UAGAGUCA-- .((((..(((((.((((((.........)))))).)))))..))))(((((.......))........(((.............)))----.....--....))).-- ( -31.52) >consensus GGCGGAAGCGUGUGACGCGGACUCACAAUGCGUCCCAUGCGGCCGCGACGCGAGAAUAGCACAUCAUCUGCA______AAGGAGGCA____AA_AA_AUAGAGA____ .((((..(((((.((((((.........)))))).)))))..))))...((.......))................................................ (-25.79 = -25.60 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:43 2006