
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
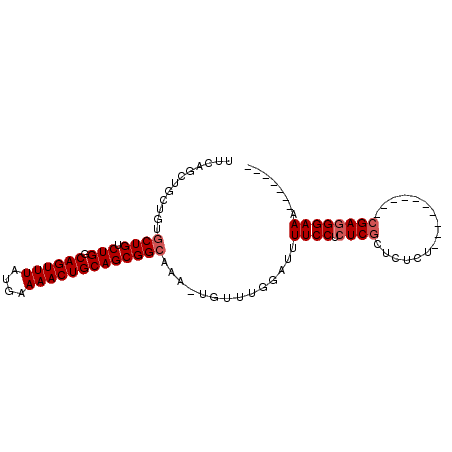
| Location | 2,193,274 – 2,193,366 |
| Length | 92 |
| Max. P | 0.988236 |

| Location | 2,193,274 – 2,193,366 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
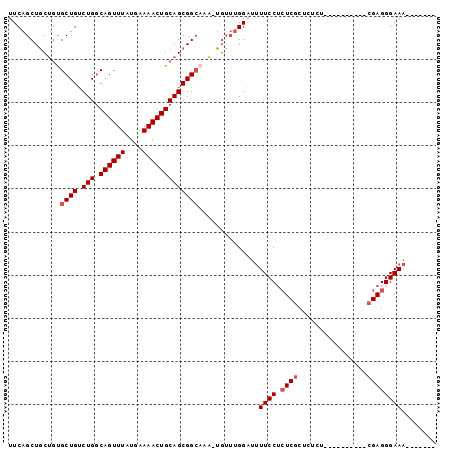
>3L_DroMel_CAF1 2193274 92 + 23771897 UUCAGCUACGAUGCUGUCUGGCAGUUUAUGAAAACUGCAGCGGCUGUUUGUUUGGAUUUUCCUCUCUCUCUCUCUCUCUCGCACGAGGGAAA------- (((((..((((.(((((...(((((((....))))))).)))))...)))))))))..................((((((....))))))..------- ( -25.60) >DroSec_CAF1 245221 77 + 1 UUCAGCUGCUGUGCUGUCUGGCAGUUUAUGAAAACUGCAGCGGAAAA-U----GGAUUUUCCUCUCGCUCUCU----------CGAGGGAAA------- ((((.((((((((((....)))(((((....))))))))))))....-)----))).(((((.((((......----------)))))))))------- ( -25.10) >DroSim_CAF1 245718 81 + 1 UUCAGCUGCUGUGCUGUCUGGCAGUUUAUGAAAACUGCAGCGGCAAA-UGUUUGGAUUUUCCUCUCGCUCUCU----------CGAGGGAAA------- (((((..((..((((((...(((((((....))))))).))))))..-.))))))).(((((.((((......----------)))))))))------- ( -27.50) >DroEre_CAF1 250494 77 + 1 UUCAGCUGCUGCGCUGUCUGCCAGUUUAUGAAAACUGCAGCGGCAAA-UGUUUGGCUCUUCC--UCGCU--CU----------CGAGGGAAC------- ....((((((((((.....)).(((((....)))))))))))))...-..........((((--(((..--..----------)))))))..------- ( -27.60) >DroYak_CAF1 244149 86 + 1 UUCAGCUGCUGUGCUGGCUGGCAGUUUAUGAAAACUGCAGCGGCAAA-UGUUUGGCUUUUCCUCUCGCU--CU----------CGAGGGAAAGCCAGGG ....(((((((((((....)))(((((....)))))))))))))...-..((((((((((((((.....--..----------.)))))))))))))). ( -39.20) >consensus UUCAGCUGCUGUGCUGUCUGGCAGUUUAUGAAAACUGCAGCGGCAAA_UGUUUGGAUUUUCCUCUCGCUCUCU__________CGAGGGAAA_______ ............((((.(((.((((((....)))))))))))))..............((((.((((................))))))))........ (-18.75 = -19.35 + 0.60)

| Location | 2,193,274 – 2,193,366 |
|---|---|
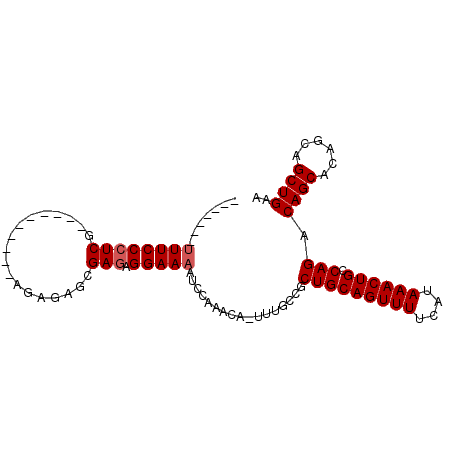
| Length | 92 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -18.67 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
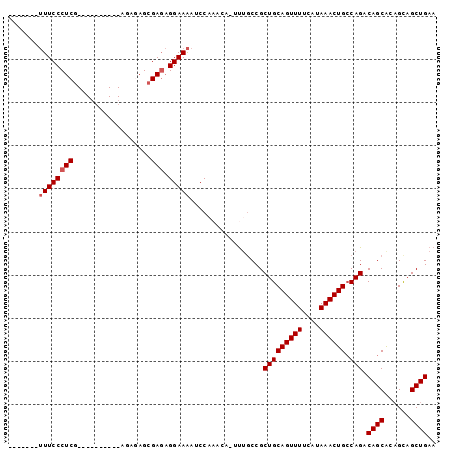
>3L_DroMel_CAF1 2193274 92 - 23771897 -------UUUCCCUCGUGCGAGAGAGAGAGAGAGAGAGGAAAAUCCAAACAAACAGCCGCUGCAGUUUUCAUAAACUGCCAGACAGCAUCGUAGCUGAA -------((((.(((...(....)...))).))))..((.....)).......((((..(.(((((((....)))))))..((.....)))..)))).. ( -23.70) >DroSec_CAF1 245221 77 - 1 -------UUUCCCUCG----------AGAGAGCGAGAGGAAAAUCC----A-UUUUCCGCUGCAGUUUUCAUAAACUGCCAGACAGCACAGCAGCUGAA -------((((.(((.----------...))).))))((((((...----.-)))))).(((((((((....)))))).))).((((......)))).. ( -26.00) >DroSim_CAF1 245718 81 - 1 -------UUUCCCUCG----------AGAGAGCGAGAGGAAAAUCCAAACA-UUUGCCGCUGCAGUUUUCAUAAACUGCCAGACAGCACAGCAGCUGAA -------(((((((((----------......)))).))))).........-.......(((((((((....)))))).))).((((......)))).. ( -23.90) >DroEre_CAF1 250494 77 - 1 -------GUUCCCUCG----------AG--AGCGA--GGAAGAGCCAAACA-UUUGCCGCUGCAGUUUUCAUAAACUGGCAGACAGCGCAGCAGCUGAA -------(((((((((----------..--..)))--))..))))......-...((.((((((((((....))))).((.....))))))).)).... ( -24.80) >DroYak_CAF1 244149 86 - 1 CCCUGGCUUUCCCUCG----------AG--AGCGAGAGGAAAAGCCAAACA-UUUGCCGCUGCAGUUUUCAUAAACUGCCAGCCAGCACAGCAGCUGAA ...(((((((((((((----------..--..)))).)).)))))))....-......((((((((((....)))))).))))((((......)))).. ( -33.90) >consensus _______UUUCCCUCG__________AGAGAGCGAGAGGAAAAUCCAAACA_UUUGCCGCUGCAGUUUUCAUAAACUGCCAGACAGCACAGCAGCUGAA .......((((((((..................))).))))).................(((((((((....)))))).))).((((......)))).. (-18.67 = -19.07 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:25 2006