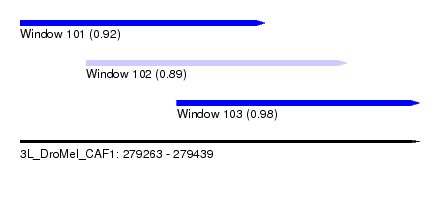
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 279,263 – 279,439 |
| Length | 176 |
| Max. P | 0.983184 |

| Location | 279,263 – 279,371 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 279263 108 + 23771897 GUAUACAUAUCGGUAAACUUUCAAUU-----------AGUUUGGUUGGGACUUAAAUUCCGGAAGAAAUGUGUGUGUAUUUGCGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCU ..((((((.((..((((((.......-----------)))))).((((((......))))))..)).))))))((((....)))).(((((((((....)))))))))((((...)))) ( -29.60) >DroSec_CAF1 51403 107 + 1 GCAUACGUCUCGAUAAACU-UCAAUU-----------AGUUUGCUUGGGACUUAAAUUCCGGAAGAAAUGUGUCUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCU ((((..(((((((((((((-......-----------)))))).)))))))........(....)..))))((((((....))...(((((((((....)))))))))..))))..... ( -32.40) >DroSim_CAF1 47198 107 + 1 GCAUACGUCUCGAUAAACU-UCAAUU-----------AGUUUACUUGGGACUUAAAUUCCGGAAGAGAUGUGUCUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCU ......(((((((((((((-......-----------)))))).)))))))............(((((.((.(((((....))...(((((((((....)))))))))))).))))))) ( -35.70) >DroEre_CAF1 50899 106 + 1 GCAUACGGAUGGAUAAACU-UCAACU------------CUUUGGUGGGGAAUUUAAUUCCGGAAGAUAUGAGUCUAUCUUUACGCAAAUUGCAGCGAAAGCUGUAAUUACAGACUCUCU ((.((.((((((((.....-(((..(------------((((.(.(((........)))).)))))..))))))))))).)).)).(((((((((....)))))))))........... ( -31.10) >DroYak_CAF1 47557 118 + 1 GCAUACGGAUCGAUCAACU-UUACUUUACUUUUUCUUACUUUGGUUGGGACUUAAAUUCCGGAAGAUAUGUGUCUAUUUUUACGCAAUUUGCAGCGAAAGCUGUAAUUACAGACUCUCU ((...((((....((((((-......................)))))).........))))(((((((......)))))))..))...(((((((....)))))))............. ( -23.67) >consensus GCAUACGUAUCGAUAAACU_UCAAUU___________AGUUUGGUUGGGACUUAAAUUCCGGAAGAAAUGUGUCUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCU ..............................................((((......))))(..((...(((((........)))))(((((((((....))))))))).....))..). (-20.32 = -20.56 + 0.24)

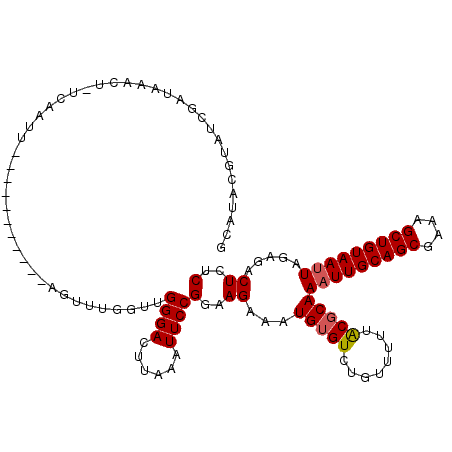
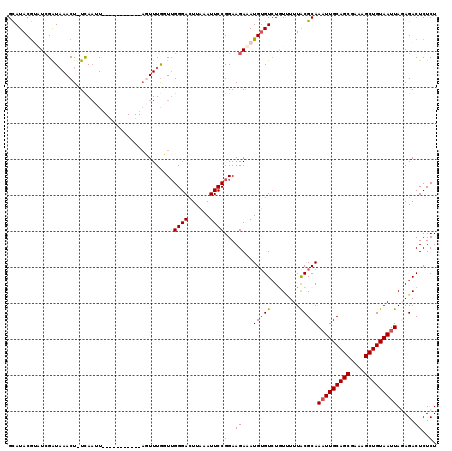
| Location | 279,292 – 279,407 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.885002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 279292 115 + 23771897 UUGGUUGGGACUUAAAUUCCGGAAGAAAUGUGUGUGUAUUUGCGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUGAUACCAAUACAAGUCAAAAUGGCAUUUAUAAU (((((.((((......))))(..(((..((((((......))))))(((((((((....)))))))))......)))..).....))))).....(((.....)))......... ( -29.90) >DroSec_CAF1 51431 115 + 1 UUGCUUGGGACUUAAAUUCCGGAAGAAAUGUGUCUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAU .((((...(((((......(....)...(((((........)))))(((((((((....)))))))))((((.....))))............))))).....))))........ ( -29.60) >DroSim_CAF1 47226 115 + 1 UUACUUGGGACUUAAAUUCCGGAAGAGAUGUGUCUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAU ..((((((((.......)))((((((((.(.((((((....))...(((((((((....)))))))))..)))).)))))))....))....))))).................. ( -31.60) >DroEre_CAF1 50926 115 + 1 UUGGUGGGGAAUUUAAUUCCGGAAGAUAUGAGUCUAUCUUUACGCAAAUUGCAGCGAAAGCUGUAAUUACAGACUCUCUCUUAAUACCAAUACACGUCAAUAUGUUAUUUGUAAU ((((((.(((((...)))))..((((...((((((...........(((((((((....)))))))))..))))))..))))..))))))......................... ( -30.12) >DroYak_CAF1 47596 115 + 1 UUGGUUGGGACUUAAAUUCCGGAAGAUAUGUGUCUAUUUUUACGCAAUUUGCAGCGAAAGCUGUAAUUACAGACUCUCUCUUAAUACCCAUACACGUCAAUAGGUUAUUCAUAAU .((..((((..........(....)...(((((........)))))..(((((((....)))))))....................))))..))..................... ( -22.90) >consensus UUGGUUGGGACUUAAAUUCCGGAAGAAAUGUGUCUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAU ......((((......))))(..(((..(((((........)))))(((((((((....)))))))))......)))..)................................... (-21.44 = -21.68 + 0.24)

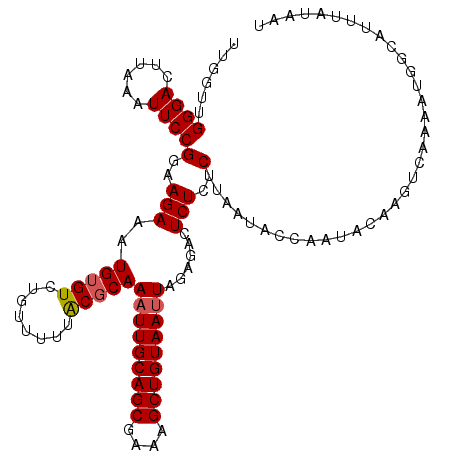
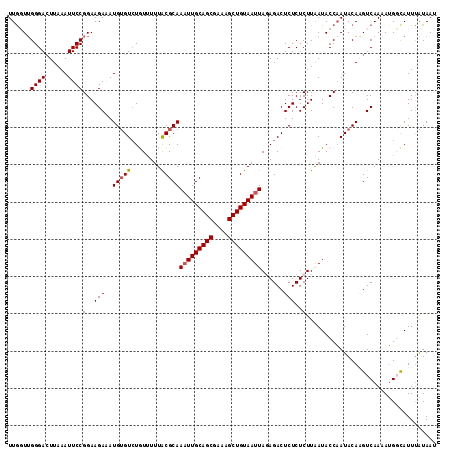
| Location | 279,332 – 279,439 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 279332 107 + 23771897 UGCGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUGAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAGUACCCCACAACGUCAUUUAAUAU ......(((((((((....)))))))))((((.....))))((((..........(((.....)))........((.((((...)))).))....))))........ ( -23.90) >DroSec_CAF1 51471 106 + 1 UACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAGUACC-CACAACGUCAUUUAAUAU ......(((((((((....)))))))))((((.....))))..................(((((((........((.((((...))))-))....)))))))..... ( -23.30) >DroSim_CAF1 47266 106 + 1 UACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAGUACC-CACAGCGUCAUUUAAUAU .((((.(((((((((....)))))))))((((.....))))..............(((.....)))........((.((((...))))-))..)))).......... ( -25.90) >DroEre_CAF1 50966 106 + 1 UACGCAAAUUGCAGCGAAAGCUGUAAUUACAGACUCUCUCUUAAUACCAAUACACGUCAAUAUGUUAUUUGUAAUGAGGCAAAGUACC-CACAACGUCAUUUAAUAU ...((.(((((((((....)))))))))...........((((.(((.((((.((((....)))))))).))).))))))........-.................. ( -22.90) >DroYak_CAF1 47636 106 + 1 UACGCAAUUUGCAGCGAAAGCUGUAAUUACAGACUCUCUCUUAAUACCCAUACACGUCAAUAGGUUAUUCAUAAUGAGGCAAAGUACC-CACAACGUCAUUUAAUAU ...((...(((((((....))))))).......(((......((((.((.............)).))))......)))))........-.................. ( -18.22) >consensus UACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAGUACC_CACAACGUCAUUUAAUAU ......(((((((((....)))))))))...(((((((....(((.(((.............))).)))......))))....((........)))))......... (-18.24 = -18.20 + -0.04)

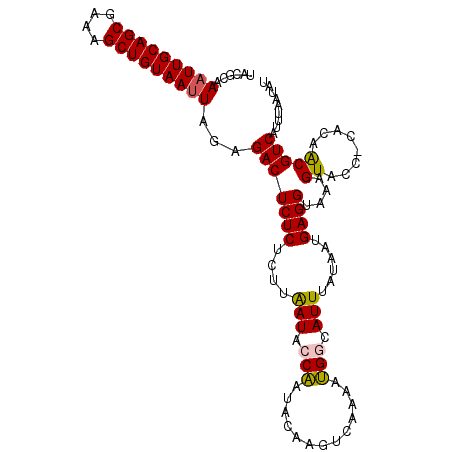
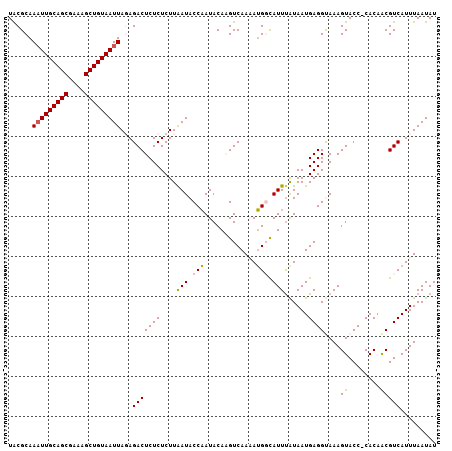
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:37 2006