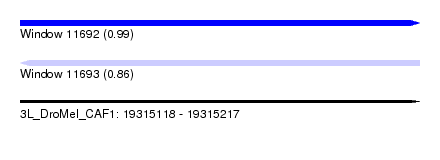
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,315,118 – 19,315,217 |
| Length | 99 |
| Max. P | 0.989881 |

| Location | 19,315,118 – 19,315,217 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.54 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.81 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
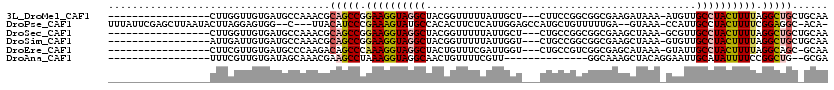
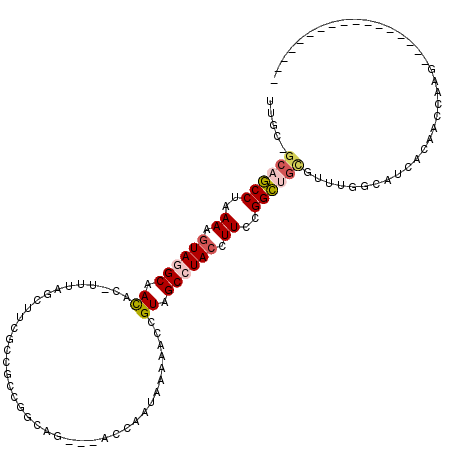
>3L_DroMel_CAF1 19315118 99 + 23771897 -----------------CUUGGUUGUGAUGCCAAACGCAGCCGGAAGGUAGGCUACGGUUUUUAUUGCU---CUUCCGGCGGCGAAGAUAAA-AUGUUGCCUACUUUUAGGCUGCUGCAA -----------------.(((((......)))))..((((((.((((((((((.((((((((..(((((---.....)))))..)))))...-.))).)))))))))).))))))..... ( -40.80) >DroPse_CAF1 85753 110 + 1 UUUAUUCGAGCUUAAUACUUAGGAGUGG--C---UUACAUCCCGAAAGUAUGCCACACUUCUCAUUGGAGCCAUGCUGUUUUUGA--GUAAA-CCAUUGCCUACUUUUCGGAGGC-ACA- ((((((((((...((((....((.((((--(---.(((.........))).))))).((((.....))))))....)))))))))--)))))-....(((((.((....))))))-)..- ( -25.80) >DroSec_CAF1 148888 99 + 1 -----------------CUUGGUUGUGAUGCCAAACGCAGCCGGAAGGUAGGCUACGGUUUUUAUUGCU---CUGCCGGCGGCGAAGCUAAA-GCGUUGCCUACUUUUAGGCUGCUGCAA -----------------.(((((......)))))..((((((.((((((((((.(((.(((.....(((---.((((...)))).))).)))-.))).)))))))))).))))))..... ( -45.70) >DroSim_CAF1 165554 99 + 1 -----------------AUUGAUUGUGAUGCCAAACGCAGCCGGAAGGUAGGCUACGGUUUUUAUUGGU---CUGCCGGCGGCGAAGCUAAA-GUGUUGCCUACUUUUAGGCUGCUGCAA -----------------...........(((.....((((((.((((((((((.(((.(((...((.(.---(((....)))).))...)))-.))).)))))))))).)))))).))). ( -37.40) >DroEre_CAF1 148622 98 + 1 -----------------CUUCGUUGUGAUGCCCAAGACAGCCCAAAGGUAGGCUACUGUUUCGAUUGGU---CUGCCGUCGGCGAGCAUAAA-GUAUUGCCUACUUUUAGGCAGC-GCAA -----------------.....(((((.((((.((((.........(((((((((..........))))---)))))((.(((((((.....-)).))))).)))))).)))).)-)))) ( -30.30) >DroAna_CAF1 131515 87 + 1 -----------------UUUCGUUGUGAUAGCAAACGAAGCCUAAAGGUAGGCAACUGUUUUCGUU--------------GGCAAAGCUACAGGAAUUGCAUAUUUUCCGGCUG--GCGA -----------------.((((((((....)).))))))(((((....)))))........((((.--------------.((.........((((.........)))).))..--)))) ( -23.90) >consensus _________________CUUGGUUGUGAUGCCAAACGCAGCCGGAAGGUAGGCUACGGUUUUUAUUGGU___CUGCCGGCGGCGAAGCUAAA_GUAUUGCCUACUUUUAGGCUGC_GCAA .....................................(((((.((((((((((.............................................)))))))))).)))))...... (-16.48 = -16.81 + 0.34)
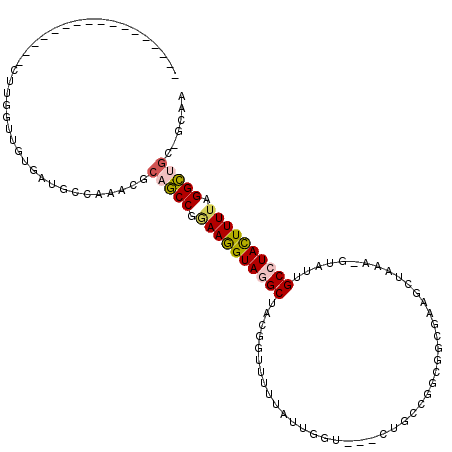
| Location | 19,315,118 – 19,315,217 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.54 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19315118 99 - 23771897 UUGCAGCAGCCUAAAAGUAGGCAACAU-UUUAUCUUCGCCGCCGGAAG---AGCAAUAAAAACCGUAGCCUACCUUCCGGCUGCGUUUGGCAUCACAACCAAG----------------- .....((((((..((.((((((.((.(-(((((((((.......))))---)....)))))...)).)))))).))..))))))..((((........)))).----------------- ( -29.60) >DroPse_CAF1 85753 110 - 1 -UGU-GCCUCCGAAAAGUAGGCAAUGG-UUUAC--UCAAAAACAGCAUGGCUCCAAUGAGAAGUGUGGCAUACUUUCGGGAUGUAA---G--CCACUCCUAAGUAUUAAGCUCGAAUAAA -(((-(((.......(((((((....)-)))))--)........((((..(((....)))..))))))))))..((((((...(((---.--..(((....))).)))..)))))).... ( -26.10) >DroSec_CAF1 148888 99 - 1 UUGCAGCAGCCUAAAAGUAGGCAACGC-UUUAGCUUCGCCGCCGGCAG---AGCAAUAAAAACCGUAGCCUACCUUCCGGCUGCGUUUGGCAUCACAACCAAG----------------- .....((((((..((.((((((.(((.-(((((((..(((...)))..---)))..))))...))).)))))).))..))))))..((((........)))).----------------- ( -35.80) >DroSim_CAF1 165554 99 - 1 UUGCAGCAGCCUAAAAGUAGGCAACAC-UUUAGCUUCGCCGCCGGCAG---ACCAAUAAAAACCGUAGCCUACCUUCCGGCUGCGUUUGGCAUCACAAUCAAU----------------- .(((.((((((..((.((((((.((..-....((......)).((...---.))..........)).)))))).))..)))))).....)))...........----------------- ( -28.50) >DroEre_CAF1 148622 98 - 1 UUGC-GCUGCCUAAAAGUAGGCAAUAC-UUUAUGCUCGCCGACGGCAG---ACCAAUCGAAACAGUAGCCUACCUUUGGGCUGUCUUGGGCAUCACAACGAAG----------------- .(((-.(.(((((((.((((((..(((-(...(((.((....)))))(---(....)).....)))))))))).)))))))......).)))...........----------------- ( -26.90) >DroAna_CAF1 131515 87 - 1 UCGC--CAGCCGGAAAAUAUGCAAUUCCUGUAGCUUUGCC--------------AACGAAAACAGUUGCCUACCUUUAGGCUUCGUUUGCUAUCACAACGAAA----------------- ((((--..((.((((.........)))).)).))...((.--------------((((((.......(((((....))))))))))).)).........))..----------------- ( -16.61) >consensus UUGC_GCAGCCUAAAAGUAGGCAACAC_UUUAGCUUCGCCGCCGGCAG___ACCAAUAAAAACCGUAGCCUACCUUCCGGCUGCGUUUGGCAUCACAACCAAG_________________ .....((((((..((.((((((.((.......................................)).)))))).))..)))))).................................... (-14.22 = -14.70 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:39 2006