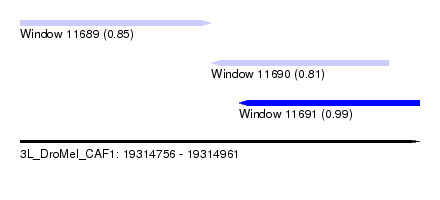
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,314,756 – 19,314,961 |
| Length | 205 |
| Max. P | 0.991518 |

| Location | 19,314,756 – 19,314,854 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19314756 98 + 23771897 GUUAUUUGUGCACGUCUGGCGAUGCAGUCGAGUGUUUAAUUUGAUUAAUUUUGUCAAGUUUAAAGCAAACACGAAUCGCUCUGCACUCUGCCAAAAAA---------------------- ................(((((((((((.(((((((((((((((((.......))))))))......))))))...)))..))))).)).)))).....---------------------- ( -24.50) >DroGri_CAF1 119202 92 + 1 GUUAUUUGUGCACGUCUGGCGAUGCACUCGAGUGUUUAAUUUGAUUAAUUUUGUCAAGCUUAAAGCAAAUACAAGU----UUGCACAGUGCCAAAU------------------------ ...(((((.(((((..((((((.....((((((.....))))))......))))))..).....((((((....))----))))...)))))))))------------------------ ( -25.30) >DroWil_CAF1 65856 92 + 1 GUUAUUUGUGCACGUCUGGCGAAGCAGUCGAGUGUUUAAUUUGAUUAAUUUUGUCAAGUCUAGAGUAAAUACGAAAUUCACUGCAUUCUGUU---------------------------- .......(((((((.(((......))).)).((((((((((((((.......)))))))......))))))).........)))))......---------------------------- ( -17.10) >DroMoj_CAF1 108817 92 + 1 GUUAUUUGUGCACGUCUGGCGAUGCAGUCGAGUGUUUAAUUUGAUUAAUUUUGUCAAGUUUUAGGCAAAUACAAAU----UUGCACUGUGCCAAAU------------------------ ...(((((.((((...((((((...((((((((.....))))))))....))))))......((((((((....))----)))).)))))))))))------------------------ ( -25.00) >DroAna_CAF1 131110 120 + 1 GUUAUUUGUGCACGUCUGGCGAUGCAGUCGAGUGUUUAAUUUGAUUAAUUUUGUCAAGUUUAAAGCAAACACGAAGCGUUCUGCACUCUGCCAAAAAAAAAAAAGAAGAAGGAAAGGUGG ................(((((((((((.((.((((((((((((((.......))))))))......))))))....))..))))).)).))))........................... ( -24.80) >DroPer_CAF1 84918 100 + 1 GUUAUUUGCGCACGUCUGGCAGUGCAGUCGAGUGUUUAAUUUGAUUAAUUUUGUCAAGUUUAAAGCAAAUACGAAAUGCUCUGCAUACUGCCAAAAAAAG-------------------- ....((((.(((.((...((((.(((.(((..(((((((((((((.......))))))))..)))))....)))..))).))))..))))))))).....-------------------- ( -27.40) >consensus GUUAUUUGUGCACGUCUGGCGAUGCAGUCGAGUGUUUAAUUUGAUUAAUUUUGUCAAGUUUAAAGCAAAUACGAAU_GCUCUGCACUCUGCCAAAA________________________ .......(((((((.(((......))).)).((((((((((((((.......))))))))......)))))).........))))).................................. (-17.03 = -17.25 + 0.22)



| Location | 19,314,854 – 19,314,945 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.79 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -25.09 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19314854 91 - 23771897 CCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGAAACUGAAGAGCUGCAAACUGGCAGCCGGAAGCUGGGAA ((((.((((((..(((....)))...................((((((.............)))))).........))))))...)))).. ( -27.02) >DroSec_CAF1 148072 90 - 1 CCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGA-ACUGAAGAGCUGCAAACUGGCAGCCGGAAGCUGGGAA ((((.((((((..(((....)))...................((((((.....-.......)))))).........))))))...)))).. ( -27.10) >DroSim_CAF1 164652 90 - 1 CCCAAUUUGGCACGCCCAAGGACCCUCACAAAUAAAUAAAACGCAGCUGGAGA-ACUGAAGAGCUGCAAACUGGCAGCCGGAAGCUGGGAA ((((.((((((..(((..((....))................((((((.....-.......)))))).....))).))))))...)))).. ( -24.70) >DroEre_CAF1 148352 90 - 1 CCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGA-ACUGGAGAGCUGCAAACUGGGAGCCGGAAGCUGGGAA ((((.((((((..(((....)))...................((((((.....-.......)))))).........))))))...)))).. ( -27.10) >consensus CCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGA_ACUGAAGAGCUGCAAACUGGCAGCCGGAAGCUGGGAA ((((.((((((..(((....)))...................((((((.............)))))).........))))))...)))).. (-25.09 = -25.34 + 0.25)


| Location | 19,314,868 – 19,314,961 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -18.31 |
| Energy contribution | -19.86 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19314868 93 - 23771897 CACCG--------------AGUGCAGAGUGCCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGAAACUGAAGAGCUGCAAACUGGCAG ..(((--------------.(((.((.(((((......)))))(((....))).))))).............((((((.............))))))....)))... ( -27.52) >DroSec_CAF1 148086 88 - 1 ----G--------------AGGGCCGAGUGCCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGA-ACUGAAGAGCUGCAAACUGGCAG ----(--------------((((((..(((((......))))).......)))))))...............((((((.....-.......)))))).......... ( -31.20) >DroSim_CAF1 164666 88 - 1 ----G--------------AGUGCCGAGUGCCCAAUUUGGCACGCCCAAGGACCCUCACAAAUAAAUAAAACGCAGCUGGAGA-ACUGAAGAGCUGCAAACUGGCAG ----.--------------.(((((((((.....)))))))))(((..((....))................((((((.....-.......)))))).....))).. ( -26.70) >DroEre_CAF1 148366 106 - 1 CACCGAGUGUAGAGUGUAGAGUGCCGAGUGCCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGA-ACUGGAGAGCUGCAAACUGGGAG ..((.(((.....(((.((.(((((((((.....)))))))))(((....))).))))).............((((((.....-.......))))))..))).)).. ( -37.40) >DroAna_CAF1 131260 72 - 1 CUCUG--------------AGUGCCGAGUGCCCAAUUUGCCACGCCCAAGGGCUCUCACAA-CAAAUACAAAAA--------------------UGCCAAAAUACAA ...((--------------((((.(((((.....))))).)))(((....)))..)))...-............--------------------............. ( -12.20) >consensus C_C_G______________AGUGCCGAGUGCCCAAUUUGGCACGCCCAAGGGCCCUCACAAAUAAAUAAAACGCAGCUGGAGA_ACUGAAGAGCUGCAAACUGGCAG ....................(((((((((.....)))))))))(((....)))...................((((((.............)))))).......... (-18.31 = -19.86 + 1.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:37 2006