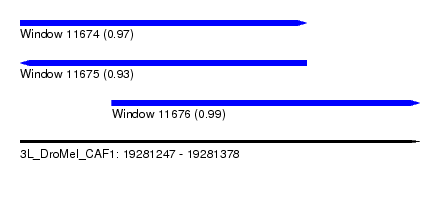
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,281,247 – 19,281,378 |
| Length | 131 |
| Max. P | 0.992711 |

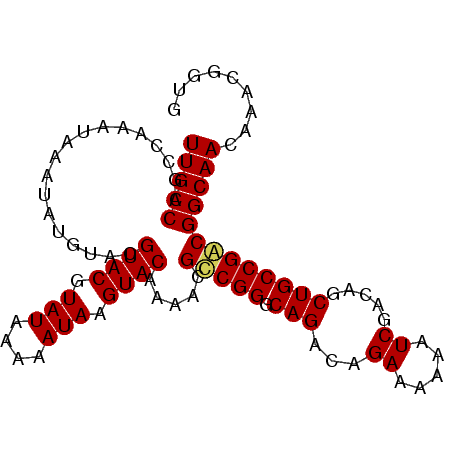
| Location | 19,281,247 – 19,281,341 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19281247 94 + 23771897 UUGCCAGCCAAAUA--UAUAUAUGUACGUAUAAAAAUAAGUACAAAACGUCGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUG (((((.........--......(((((.(((....))).)))))....(((((.(((...((.....)).....)))))))))))))......... ( -23.80) >DroSec_CAF1 114623 96 + 1 UUGCCAGCCAAAUAAAUAUGUACGUACGUAUAAAAAUAAGUACAAAACGCCGGGCAGACAGAAAAAAUCGACAGCUGCCGGCGGCAACAAAUGGUG ..(((((((..............((((.(((....))).)))).....(((((.(((...((.....)).....)))))))))))......)))). ( -25.60) >DroSim_CAF1 121426 96 + 1 UUGCCAGCCAAAUAAAUAUGUACGUACGUAUAAAAAUAAGUACAAAACGCCGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUG ......(((..............((((.(((....))).)))).....(((((((((...((.....)).....)))))..)))).......))). ( -22.80) >consensus UUGCCAGCCAAAUAAAUAUGUACGUACGUAUAAAAAUAAGUACAAAACGCCGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUG (((((..................((((.(((....))).)))).....(((((.(((...((.....)).....)))))))))))))......... (-22.05 = -22.17 + 0.11)


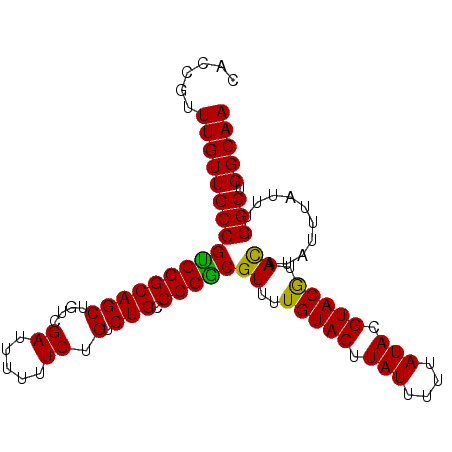
| Location | 19,281,247 – 19,281,341 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -24.05 |
| Energy contribution | -23.17 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19281247 94 - 23771897 CACCGUUUGUUGCCGUCGGCAGCUGUCGAUUUUUUCUGUCUGCCCGACGUUUUGUACUUAUUUUUAUACGUACAUAUAUA--UAUUUGGCUGGCAA ...((...((((((...))))))...))........((((.(((.((..(..(((((.(((....))).))))).....)--..)).))).)))). ( -22.90) >DroSec_CAF1 114623 96 - 1 CACCAUUUGUUGCCGCCGGCAGCUGUCGAUUUUUUCUGUCUGCCCGGCGUUUUGUACUUAUUUUUAUACGUACGUACAUAUUUAUUUGGCUGGCAA ......(((((((((((((((((....((.....)).).))).)))))((..(((((.(((....))).))))).))..........))).))))) ( -25.30) >DroSim_CAF1 121426 96 - 1 CACCGUUUGUUGCCGUCGGCAGCUGUCGAUUUUUUCUGUCUGCCCGGCGUUUUGUACUUAUUUUUAUACGUACGUACAUAUUUAUUUGGCUGGCAA ......(((((((((((((((((....((.....)).).))).)))))((..(((((.(((....))).))))).))..........))).))))) ( -22.60) >consensus CACCGUUUGUUGCCGUCGGCAGCUGUCGAUUUUUUCUGUCUGCCCGGCGUUUUGUACUUAUUUUUAUACGUACGUACAUAUUUAUUUGGCUGGCAA ......(((((((((((((((((....((.....)).).))).)))))((..(((((.(((....))).))))).))..........))).))))) (-24.05 = -23.17 + -0.88)

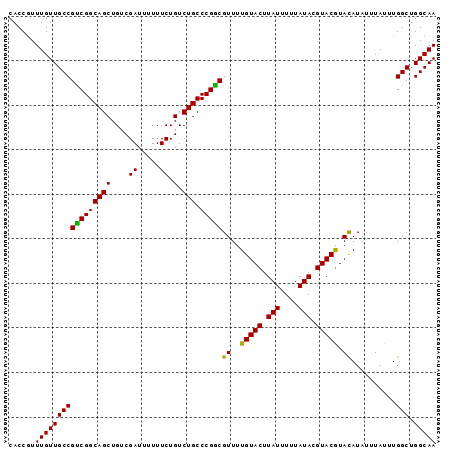
| Location | 19,281,277 – 19,281,378 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19281277 101 + 23771897 AAAAUAAGUACAAAAC-GUCGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAG ................-(((.....)))........(((((((((((((..(....)...))))((((((((....)))))))).......))).)))))). ( -27.10) >DroSec_CAF1 114655 101 + 1 AAAAUAAGUACAAAAC-GCCGGGCAGACAGAAAAAAUCGACAGCUGCCGGCGGCAACAAAUGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAG ...............(-(((((.(((...((.....)).....))))))))).(((((........((((((....))))))((((....)))).))))).. ( -28.90) >DroSim_CAF1 121458 101 + 1 AAAAUAAGUACAAAAC-GCCGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAG ....(((((.....((-(((((((((...((.....)).....)))))...(....)...))))))((((((....)))))))))))............... ( -26.90) >DroEre_CAF1 114258 100 + 1 AAAAUAAGUCCAAAAA-GGCAGGCAGACAGAA-AAAUCGACAUCUGCCAACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAG .......(((......-))).((((((..((.-...))....)))))).....(((((..((..((((((((....)))))))).....))....))))).. ( -23.90) >DroYak_CAF1 146941 101 + 1 AAAAUAAGUAAAAAAAAGCCGGGCAGACAGAA-AAAUCGACAUCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAG ..((((.((........((((((((((..((.-...))....))))))..))))......((..((((((((....)))))))).....)).)).))))... ( -26.50) >consensus AAAAUAAGUACAAAAC_GCCGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAG ..((((.((........(((((((((...((.....)).....)))))..))))......((..((((((((....)))))))).....)).)).))))... (-21.90 = -22.02 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:23 2006