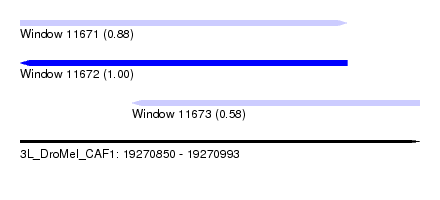
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,270,850 – 19,270,993 |
| Length | 143 |
| Max. P | 0.999721 |

| Location | 19,270,850 – 19,270,967 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.03 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -9.50 |
| Energy contribution | -12.90 |
| Covariance contribution | 3.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19270850 117 + 23771897 UAAAGAAAGUUAAGAAGAGAGAACAUCCGAAAUCAAGAAUGUAGCGGUUUUUCUAUGUAUUCCACCCAUCUCUGGAAUACGUUGAUUACCAAGUCUUUCUUGAUCCUGAGGAUAUUC ....................(((.((((...((((((((.(....(((...((.((((((((((........)))))))))).))..)))....).)))))))).....)))).))) ( -30.10) >DroSec_CAF1 99637 117 + 1 UGAAGAGUAUUAAGAAGAUAUAACAUCCAAAAUCAAGAAUCUACUGGUUUUAACAUAUAUUCCAGCAAUCUCCGGAAUACGUUGAUUACCAAGUCUUUCUUGAUCCUGAAGAUAUCC ................(((((......((..((((((((...((((((.(((((...((((((..........)))))).)))))..))).)))..))))))))..))...))))). ( -23.60) >DroSim_CAF1 111066 110 + 1 UAAG-------AAGCAGAGAAGUCAUCCAAAAUCAAGAAUCUAGUGGUUUCAACAUGUAUUCCAGCAAUUUCCGGAAUACGUUGAUUACCAAGUCUUUCUUGAUCCUAAUGAUAUCC ....-------.......((.(((((.....((((((((.....((((.(((((..(((((((..........))))))))))))..)))).....))))))))....))))).)). ( -30.10) >DroEre_CAF1 103930 90 + 1 UAAAGGAAGUUAAGAAGAGAGAAUAUCCAAAAUCAAGAAUCUAGUGGGUUUUCUAUGUAUUCCG---------------------------AGUAUUUCUUGAUCUAACAGAUAUUC ....................(((((((....((((((((.....((((............))))---------------------------.....))))))))......))))))) ( -15.60) >DroYak_CAF1 129718 117 + 1 UAAAGGAAGUUGAGAAGAGAAAAUGUCCAACAUUAAGAAUCUACUGGUUUUUCAAUGUAUUCGACGCCUUUCGUAUAUACGCUGAUUACCAAGUAUUUCUUUAUCUUGAAGAUAUUC .........(..(((((((((((((.....)))).......(((((((...(((.(((((.(((......)))...))))).)))..))).)))).)))))).)))..)........ ( -22.80) >consensus UAAAGAAAGUUAAGAAGAGAAAACAUCCAAAAUCAAGAAUCUAGUGGUUUUUCCAUGUAUUCCACCAAUCUCCGGAAUACGUUGAUUACCAAGUCUUUCUUGAUCCUGAAGAUAUUC ........................(((....((((((((.....((((......(((((((((..........))))))))).....)))).....))))))))......))).... ( -9.50 = -12.90 + 3.40)


| Location | 19,270,850 – 19,270,967 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.03 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -16.98 |
| Energy contribution | -19.58 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.60 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19270850 117 - 23771897 GAAUAUCCUCAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCAGAGAUGGGUGGAAUACAUAGAAAAACCGCUACAUUCUUGAUUUCGGAUGUUCUCUCUUCUUAACUUUCUUUA ((((((((...((((((((((.(...((((..((...((((((((........))))))))...))...))))...).)))))))))).)))))))).................... ( -31.70) >DroSec_CAF1 99637 117 - 1 GGAUAUCUUCAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCGGAGAUUGCUGGAAUAUAUGUUAAAACCAGUAGAUUCUUGAUUUUGGAUGUUAUAUCUUCUUAAUACUCUUCA ((((((.((((((((((((((..(((.(((....((((((((((((......)))))..)))))))...))))))...))))))))))))))....))))))............... ( -33.10) >DroSim_CAF1 111066 110 - 1 GGAUAUCAUUAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCGGAAAUUGCUGGAAUACAUGUUGAAACCACUAGAUUCUUGAUUUUGGAUGACUUCUCUGCUU-------CUUA (((..((((((((((((((((.....((((..((((((((((((((......)))))))))..))))).)))).....))))))))))).)))))..)))......-------.... ( -36.30) >DroEre_CAF1 103930 90 - 1 GAAUAUCUGUUAGAUCAAGAAAUACU---------------------------CGGAAUACAUAGAAAACCCACUAGAUUCUUGAUUUUGGAUAUUCUCUCUUCUUAACUUCCUUUA ((((((((...((((((((((.....---------------------------.((..............))......)))))))))).)))))))).................... ( -17.44) >DroYak_CAF1 129718 117 - 1 GAAUAUCUUCAAGAUAAAGAAAUACUUGGUAAUCAGCGUAUAUACGAAAGGCGUCGAAUACAUUGAAAAACCAGUAGAUUCUUAAUGUUGGACAUUUUCUCUUCUCAACUUCCUUUA (((.((.(((((.((.(((((.((((.(((..((((.((((..(((.....)))...)))).))))...)))))))..))))).)).))))).)).))).................. ( -22.70) >consensus GAAUAUCUUCAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCGGAAAUGGCUGGAAUACAUAGAAAAACCACUAGAUUCUUGAUUUUGGAUGUUCUCUCUUCUUAACUUCCUUUA ((((((((...((((((((((.....((((.......((((((((........))))))))........)))).....)))))))))).)))))))).................... (-16.98 = -19.58 + 2.60)



| Location | 19,270,890 – 19,270,993 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -17.64 |
| Energy contribution | -19.82 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19270890 103 - 23771897 UCUCGGUGGGAAUUGUGUUGGGAGAAGAAUAUCCUCAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCAGAGAUGGGUGGAAUACAUAGAAAAACCGCUAC ....(((((.......(((((((........))).....((((((.....))))))...))))((((((((........)))))))).........))))).. ( -27.10) >DroSec_CAF1 99677 103 - 1 UUUCGGUGGGAAUUUUGUUGGGAGAAGGAUAUCUUCAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCGGAGAUUGCUGGAAUAUAUGUUAAAACCAGUAG ....(((...(((...(((((((((......)))))...((((((.....))))))...))))(((((((((......)))))))))..)))...)))..... ( -25.80) >DroSim_CAF1 111099 103 - 1 UCUCGGUGGCAAUUGUGUUGGGAGAAGGAUAUCAUUAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCGGAAAUUGCUGGAAUACAUGUUGAAACCACUAG ....(((..((((...(((((((........))......((((((.....))))))..)))))(((((((((......)))))))))..))))..)))..... ( -28.10) >DroYak_CAF1 129758 103 - 1 UCUCGGAGGGAAUUAUAUUGCGAGAGGAAUAUCUUCAAGAUAAAGAAAUACUUGGUAAUCAGCGUAUAUACGAAAGGCGUCGAAUACAUUGAAAAACCAGUAG (((.((((......(((((.(....).))))))))).)))........((((.(((..((((.((((..(((.....)))...)))).))))...))))))). ( -19.00) >consensus UCUCGGUGGGAAUUGUGUUGGGAGAAGAAUAUCUUCAGGAUCAAGAAAGACUUGGUAAUCAACGUAUUCCGGAAAUGGCUGGAAUACAUGGAAAAACCACUAG ....(((((.......(((((..((((.....))))...((((((.....))))))..)))))(((((((((......))))))))).........))))).. (-17.64 = -19.82 + 2.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:20 2006