
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,262,583 – 19,262,715 |
| Length | 132 |
| Max. P | 0.917264 |

| Location | 19,262,583 – 19,262,696 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -43.03 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.45 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19262583 113 + 23771897 GCAUUACGAGCCAAAGCUUCCGGAUUGGUGUCGUCAGGCGGUGGAGAUAGCCGCUCUCCUUCACGACAUUGGUCACGGACCAAUGUCACAUGCCUGGGAG----UUGGUGACCCACC ((...(((((((((..(....)..))))).))))...))(((((.(...((((..(((((.((.((((((((((...))))))))))...))...)))))----.))))..)))))) ( -42.90) >DroPse_CAF1 33631 113 + 1 UAAAUAUGAGACUAAGCUGCCCGAUUGGUGUCGAAAGGCGGUAGAGAUAGCAGCUCUUCUGCACGACAUCGGUCACGGACCGAUGUCGCAUGCCUGGGAA----AAAGCUUCCCAUA ........(((...((((((....(((.((((....)))).))).....))))))..)))((((((((((((((...)))))))))))..))).((((((----.....)))))).. ( -45.20) >DroEre_CAF1 95544 113 + 1 GCACUACGAGCCAAAACUUCCGGACUGGUGUCGUCAGGCGGUGGAGAUAGCCGCCCUCCUUCACGACAUUGGACAUGGGCCAGUGUCGCACACCUGGGAG----UUGGCGAGCCAUC .........((((..((((((((....((((.....((((((.......)))))).........((((((((.......))))))))))))..)))))))----)))))........ ( -43.30) >DroMoj_CAF1 40791 113 + 1 CAACUACGAGCGAGAGCUGCCCGAAUGGUGCCGUCGAUCGGUGGAGAUUGCGGCGCUGCUGCACGAUGUCGGACAUGGUCCAUUUUCGCAUUACUGGGAA----CAAGUUUGUGGCG ...((((((((((((((.((......(((((((.(((((......)))))))))))))).))........((((...))))..)))))).....((....----))...)))))).. ( -38.80) >DroAna_CAF1 81425 113 + 1 GCACUAUGAGCCAAAACUACCGGAUUGGUGUCGCCAAGCGGUGGAAAUAGCCGCUCUACUUCACGAUAUUGGCCAUGGGCCAAUGUCACAUGCUUGGGAACGUCUUGGUGA----AC .(((((.(((((((..(....)..)))))((..(((((((((((......))))).........((((((((((...))))))))))....))))))..)).)).))))).----.. ( -42.70) >DroPer_CAF1 33727 113 + 1 UAAAUAUGAGACUAAGCUGCCCGAUUGGUGUCGAAAGGCGGUAGAGAUAGCAGCUCUCCUGCACGACAUCGGUCACGGACCGAUGUCGCAUGCCUGGGAA----AAAGCUUCCCAUA .......((((....(((((....(((.((((....)))).))).....)))))))))..((((((((((((((...)))))))))))..))).((((((----.....)))))).. ( -45.30) >consensus GAACUACGAGCCAAAGCUGCCCGAUUGGUGUCGUCAGGCGGUGGAGAUAGCCGCUCUCCUGCACGACAUCGGUCACGGACCAAUGUCGCAUGCCUGGGAA____UAAGCGACCCAUC ...............(((....(((....))).(((((((((((((.......)))).......((((((((((...)))))))))))).))))))).........)))........ (-21.64 = -21.45 + -0.19)


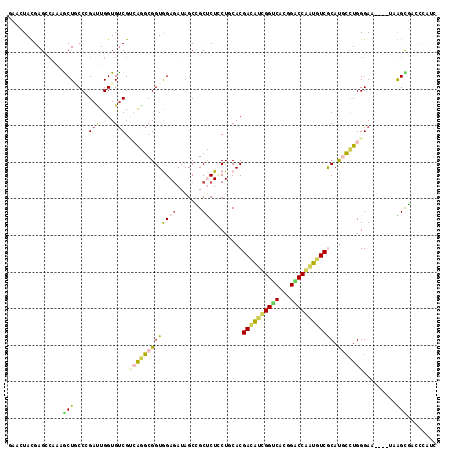
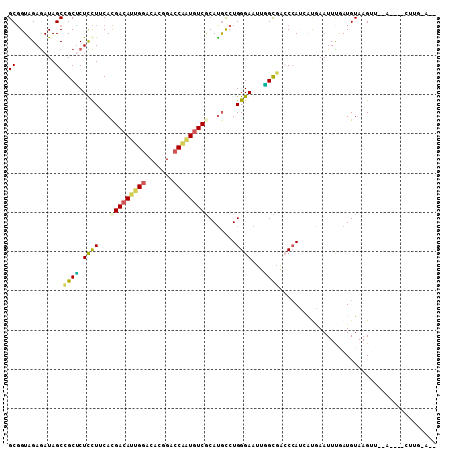
| Location | 19,262,583 – 19,262,696 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -20.05 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
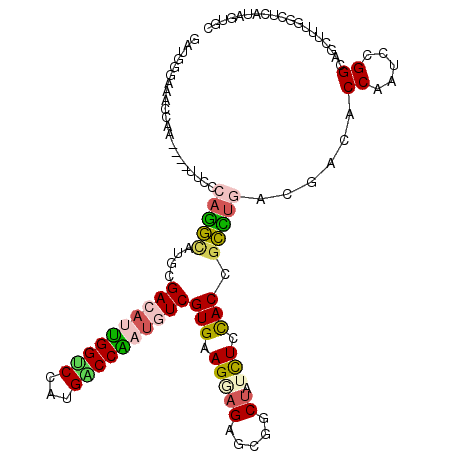
>3L_DroMel_CAF1 19262583 113 - 23771897 GGUGGGUCACCAA----CUCCCAGGCAUGUGACAUUGGUCCGUGACCAAUGUCGUGAAGGAGAGCGGCUAUCUCCACCGCCUGACGACACCAAUCCGGAAGCUUUGGCUCGUAAUGC ((((...))))..----....(((((....((((((((((...)))))))))).....(((((.......)))))...)))))((((..((((..(....)..)))).))))..... ( -45.00) >DroPse_CAF1 33631 113 - 1 UAUGGGAAGCUUU----UUCCCAGGCAUGCGACAUCGGUCCGUGACCGAUGUCGUGCAGAAGAGCUGCUAUCUCUACCGCCUUUCGACACCAAUCGGGCAGCUUAGUCUCAUAUUUA (((((((((((..----..(((((((..((((((((((((...))))))))))))((((.....))))..........))))...((......))))).))))...))))))).... ( -43.80) >DroEre_CAF1 95544 113 - 1 GAUGGCUCGCCAA----CUCCCAGGUGUGCGACACUGGCCCAUGUCCAAUGUCGUGAAGGAGGGCGGCUAUCUCCACCGCCUGACGACACCAGUCCGGAAGUUUUGGCUCGUAGUGC ((((((.((((..----((((......(((((((.((((....).))).)))))))..))))))))))))))......(((((.(((..((((..(....)..)))).)))))).)) ( -43.30) >DroMoj_CAF1 40791 113 - 1 CGCCACAAACUUG----UUCCCAGUAAUGCGAAAAUGGACCAUGUCCGACAUCGUGCAGCAGCGCCGCAAUCUCCACCGAUCGACGGCACCAUUCGGGCAGCUCUCGCUCGUAGUUG .((.((.......----(((.((....)).)))..(((((...))))).....))))..(((((((((.(((......))).).))))......(((((.......)))))..)))) ( -27.40) >DroAna_CAF1 81425 113 - 1 GU----UCACCAAGACGUUCCCAAGCAUGUGACAUUGGCCCAUGGCCAAUAUCGUGAAGUAGAGCGGCUAUUUCCACCGCUUGGCGACACCAAUCCGGUAGUUUUGGCUCAUAGUGC ..----...((((((((((.((((((....((.(((((((...))))))).))(((((((((.....)))))).))).)))))).)))(((.....))).))))))).......... ( -40.90) >DroPer_CAF1 33727 113 - 1 UAUGGGAAGCUUU----UUCCCAGGCAUGCGACAUCGGUCCGUGACCGAUGUCGUGCAGGAGAGCUGCUAUCUCUACCGCCUUUCGACACCAAUCGGGCAGCUUAGUCUCAUAUUUA (((((((((((..----..(((((((..((((((((((((...))))))))))))...((((((.......)))).))))))...((......))))).))))...))))))).... ( -44.30) >consensus GAUGGGAAACCAA____UUCCCAGGCAUGCGACAUUGGUCCAUGACCAAUGUCGUGAAGGAGAGCGGCUAUCUCCACCGCCUGACGACACCAAUCCGGCAGCUUUGGCUCAUAGUGC .....................(((((....((((((((((...))))))))))(((.(((((.....)).))).))).)))))......((.....))................... (-20.05 = -20.17 + 0.12)

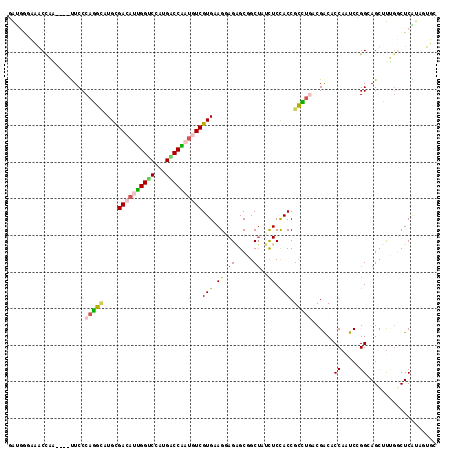
| Location | 19,262,620 – 19,262,715 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.85 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -15.07 |
| Energy contribution | -14.22 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19262620 95 + 23771897 GCGGUGGAGAUAGCCGCUCUCCUUCACGACAUUGGUCACGGACCAAUGUCACAUGCCUGGGAGUUGGUGACCCACCAUGAAUUUGAUGUAAGUU--A------------ ..(((((.(...((((..(((((.((.((((((((((...))))))))))...))...))))).))))..))))))..................--.------------ ( -34.50) >DroPse_CAF1 33668 107 + 1 GCGGUAGAGAUAGCAGCUCUUCUGCACGACAUCGGUCACGGACCGAUGUCGCAUGCCUGGGAAAAAGCUUCCCAUAACAAAUUCAAUGUAAGAUUCGUGGUCUUGGA-- ((((.((((.......)))).)))).(((((((((((...)))))))))))....((((((((.....))))))...............(((((.....))))))).-- ( -38.10) >DroSim_CAF1 102967 95 + 1 GCGGUGGAGAUAGCCGCUCUCCUUCACGACAUUGGACACGGACCAAUGUCACAUGCCUGGGAGUUGGUAACCAACCAUGAAUUUGAUGUAAGUU--A------------ (((((.......))))).(((((.((.((((((((.......))))))))...))...))))).((((.....)))).................--.------------ ( -26.40) >DroEre_CAF1 95581 103 + 1 GCGGUGGAGAUAGCCGCCCUCCUUCACGACAUUGGACAUGGGCCAGUGUCGCACACCUGGGAGUUGGCGAGCCAUCAUGAAUUUGAUGUAAGUA--A---ACCUGCA-G ((((.(......(((((((((((...(((((((((.......))))))))).......)))))..)))).))(((((......)))))......--.---.))))).-. ( -37.10) >DroWil_CAF1 11600 106 + 1 GCUGUAGAAAUAGCCGCACUUUUACAUGACAUAGGACAUGGAGCAUUUUCGCAUGUCUGGGAAUGUGUGUGCCAGGGUCAAUUUGAUGUAAGUAUAG---AAUAGAAAG (((((....))))).((((...(((((..(...((((((((((....))).))))))).)..))))).))))..(.(((.....))).)........---......... ( -26.50) >DroPer_CAF1 33764 107 + 1 GCGGUAGAGAUAGCAGCUCUCCUGCACGACAUCGGUCACGGACCGAUGUCGCAUGCCUGGGAAAAAGCUUCCCAUAACAAAUUCAAUGUAAGAUUCGUGGUCUUGGA-- ((((.((((.......)))).)))).(((((((((((...)))))))))))....((((((((.....))))))...............(((((.....))))))).-- ( -38.10) >consensus GCGGUAGAGAUAGCCGCUCUCCUUCACGACAUUGGACACGGACCAAUGUCGCAUGCCUGGGAAUUGGCGACCCAUCAUGAAUUUGAUGUAAGUU__A____CUUG_A__ ((..........))((((.((((...(((((((((.......))))))))).......))))...))))........................................ (-15.07 = -14.22 + -0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:16 2006